Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (20): 4181-4189.doi: 10.12307/2025.633
Screening and validation of glucose metabolism genes in osteoarthritis
Liu Kexin1, Ma Chao2, Liu Kai1, Hao Maochen1, Wang Xingru1, Meng Lingting1, Dong Mei3, Wang Jianzhong2
- 1Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China; 2Department of Trauma Surgery Center C, 3Department of Osteoporosis, Second Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China
-
Received:2024-06-07Accepted:2024-08-05Online:2025-07-18Published:2024-12-19 -
Contact:Wang Jianzhong, MD, Chief physician, Professor, Department of Trauma Surgery Center C, Second Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China Co-corresponding author: Dong Mei, Department of Osteoporosis, Second Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China -
About author:Liu Kexin, Master candidate, Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China Ma Chao, MS, Attending physician, Department of Trauma Surgery Center C, Second Affiliated Hospital of Inner Mongolia Medical University, Hohhot 010030, Inner Mongolia Autonomous Region, China Liu Kexin and Ma Chao contributed equally to this article. -
Supported by:Inner Mongolia Medical University Key Project, No. YKD2024ZD005 (to WJZ); Inner Mongolia Autonomous Region Third Batch of Capital Area Public Hospital High-level Clinical Specialty Construction Science and Technology Project, No. 2023SGGZ143 (to WJZ); Inner Mongolia Autonomous Region Directly Affiliated Universities Basic Scientific Research Business Expenses Project, No. YKD2023ZY001 (to LK); Inner Mongolia Autonomous Region Postgraduate Scientific Research Innovation Project, No. S20231189Z (to LK)
CLC Number:
Cite this article
Liu Kexin, Ma Chao, Liu Kai, Hao Maochen, Wang Xingru, Meng Lingting, Dong Mei, Wang Jianzhong . Screening and validation of glucose metabolism genes in osteoarthritis[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(20): 4181-4189.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
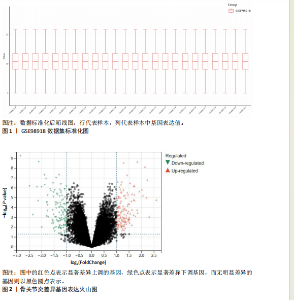
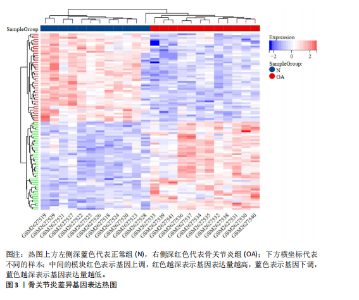
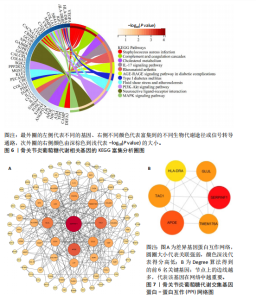
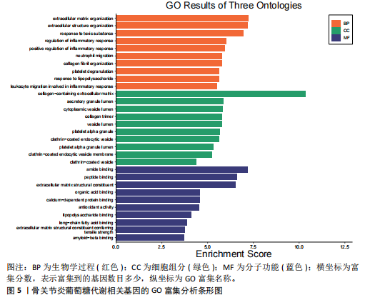
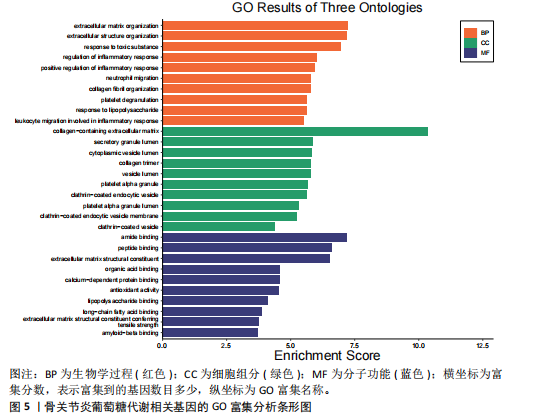
2.1 数据集获取 在GEO数据库中获取符合筛选条件的数据集GSE98918,该数据集包含12例骨关节炎患者的半月板组织和12例非骨关节炎(关节镜下半月板部分切除术)患者的半月板组织。 2.2 数据处理及差异基因筛选 运用R语言对GSE98918数据集进行差异表达基因的数据标准化预处理,标准化结果以箱式图呈现(图1)。使用R语言中的“limma”包,并且设定筛选条件为:P < 0.05且|log2FC|> 1,获得266个差异表达基因,其中上调基因145个、下调基因121个。使用“ggplot2”工具包绘制火山图(图2)及“pheatmap”工具包绘制热图(图3)。 2.3 交集基因 通过查询GeneCards数据库,得到了10 899个与葡萄糖代谢相关的基因。与之前得到的骨关节炎差异表达基因进行交集分析后,发现有134个基因与骨关节炎和葡萄糖代谢相关,绘制韦恩图(图4)。 2.4 GO和KEGG通路富集分析 对134个与骨关节炎和葡萄糖代谢相关基因进行了GO和KEGG富集分析,GO富集分析的结果显示:骨关节炎葡萄糖代谢相关基因在生物学过程中主要参与对有毒物质的反应、炎症反应的正调节、中性粒细胞迁移、血小板脱颗粒、对脂多糖的反应、参与炎症反应的白细胞迁移等过程;细胞组分显示其主要存在于含胶原蛋白的细胞外基质、分泌性颗粒腔、细胞质囊泡腔、胶原蛋白三聚体、网格蛋白包被的内吞囊泡、血小板α颗粒腔等;分子功能上,其在与酰胺结合、肽结合、构成细胞外基质结构成分、有机酸结合、钙依赖性蛋白结合、抗氧化活性、脂多糖结合、长链脂肪酸结合、淀粉样蛋白-β结合中发挥作用(图5)。 根据KEGG富集分析结果显示:骨关节炎葡萄糖代谢相关基因在磷脂酰肌醇3-激酶-蛋白激酶B (phosphatidylinositol 3 kinase-protein"
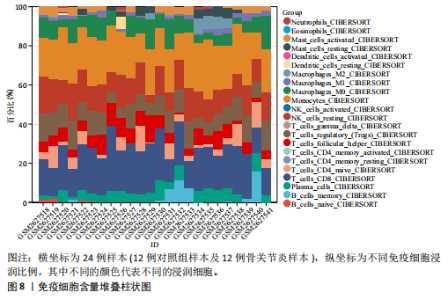
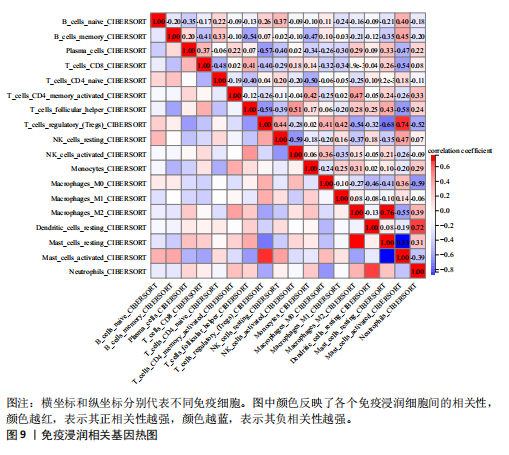
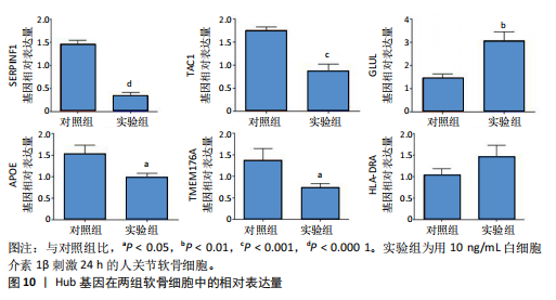
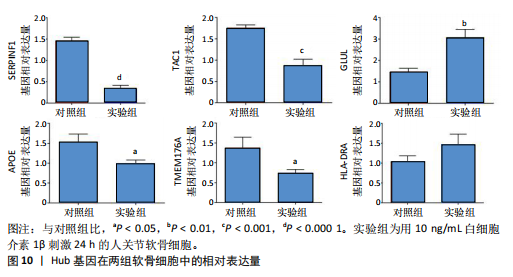
kinase B,PI3K-Akt)信号通路、金黄色葡萄球菌感染、神经活性配体-受体相互作用、补体和凝血级联反应、白细胞介素17(interleukin-17,IL-17)信号通路、类风湿性关节炎、糖尿病并发症中的晚期糖基化终末产物及其受体(advanced glycation end products-receptor for advanced glycation end products,AGE-RAGE)信号通路、流体剪切应力和动脉粥样硬化、胆固醇代谢、丝裂原活化蛋白激酶(MAPK)信号通路、1型糖尿病等中显著富集(图6)。 2.5 PPI网络分析 通过将134个交集基因输入STRING数据库,获得PPI网络图(图7A),并将结果导入Cytoscape软件,运用插件cytoHubba中Degree算法进行关键基因筛选,筛选评分最高的6个Hub基因,分别为APOE、SERPINF1、TAC1、HLA-DRA、GLUL、TMEM176A(图7B)。 2.6 免疫浸润分析 采用CIBERSORT算法对GSE98918数据集进行免疫细胞浸润情况评估和可视化,骨关节炎样本中主要免疫浸润细胞为:巨噬细胞、单核细胞、静息状态下的NK细胞、调节性T细胞、CD8+ T细胞,将免疫浸润分析结果通过免疫细胞含量堆叠柱状图展现(图8)。采用pearson法免疫细胞相关性分析,绘制免疫浸润相关基因热图(图9)。结果显示,M2型巨噬细胞与静息肥大细胞表现出最强的正相关性,而活化肥大细胞和静息肥大细胞则表现出最强的负相关性。 2.7 实验验证 如图10所示,相比于对照组,实验组中Hub基因SERPINF1的mRNA表达显著降低(P < 0.000 1),TAC1的mRNA表达显著降低(P < 0.001),GLUL的mRNA表达显著升高"
| [1] JANG S, LEE K, JU JH. Recent Updates of Diagnosis, Pathophysiology, and Treatment on Osteoarthritis of the Knee. Int J Mol Sci. 2021;22(5):2619. [2] ALLEN KD, THOMA LM, GOLIGHTLY YM. Epidemiology of osteoarthritis. Osteoarthritis Cartilage. 2022;30(2):184-195. [3] DUONG V, OO W M, DING C, et al. Evaluation and Treatment of Knee Pain: A Review. JAMA. 2023;330(16):1568-1580. [4] 钱燕,刘启颂.调整细胞培养条件提高间充质干细胞外泌体治疗骨关节炎的潜能[J].中国组织工程研究,2025,29(1):164-174. [5] YU M, XU Y, WENG X, et al. Clinical outcome and survival rate of condylar constrained knee prosthesis in revision total knee arthroplasty: an average nine point six year follow-up. Int Orthop. 2024;48(5):1179-1187. [6] HOLLANDER JM, ZENG L. The Emerging Role of Glucose Metabolism in Cartilage Development. Curr Osteoporos Rep. 2019;17(2):59-69. [7] ZLACKÁ J, ZEMAN M. Glycolysis under Circadian Control. Int J Mol Sci. 2021; 22(24):13666. [8] OHASHI Y, TAKAHASHI N, TERABE K, et al. Metabolic reprogramming in chondrocytes to promote mitochondrial respiration reduces downstream features of osteoarthritis. Sci Rep. 2021;11(1):15131. [9] KONG P, CHEN R, ZOU FQ, et al. HIF-1α repairs degenerative chondrocyte glycolytic metabolism by the transcriptional regulation of Runx2. Eur Rev Med Pharmacol Sci. 2021;25(3):1206-1214. [10] ARRUDA AL, HARTLEY A, KATSOULA G, et al. Genetic underpinning of the comorbidity between type 2 diabetes and osteoarthritis. Am J Hum Genet. 2023;110(8):1304-1318. [11] PEMMARI A, LEPPÄNEN T, HÄMÄLÄINEN M, et al. Widespread regulation of gene expression by glucocorticoids in chondrocytes from patients with osteoarthritis as determined by RNA-Seq. Arthritis Res Ther. 2020;22(1):271. [12] UESAKA K, OKA H, KATO R, et al. Bioinformatics in bioscience and bioengineering: Recent advances, applications, and perspectives. J Biosci Bioeng. 2022;134(5):363-373. [13] BARRETT T, WILHITE SE, LEDOUX P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991-995. [14] BROPHY RH, ZHANG B, CAI L, et al. Transcriptome comparison of meniscus from patients with and without osteoarthritis. Osteoarthritis Cartilage. 2018;26(3):422-432. [15] 周亮,陈兴真,李振宇,等. lncRNA HOTAIR在白细胞介素1β介导骨关节炎中的作用机制[J].中国组织工程研究,2022, 26(35):5607-5613. [16] HAWKER GA. Osteoarthritis is a serious disease. Clin Exp Rheumatol. 2019;37 Suppl 120(5):3-6. [17] LONG H, LIU Q, YIN H, et al. Prevalence Trends of Site-Specific Osteoarthritis From 1990 to 2019: Findings From the Global Burden of Disease Study 2019. Arthritis Rheumatol. 2022;74(7):1172-1183. [18] LI K, JI X, SEELEY R, et al. Impaired glucose metabolism underlies articular cartilage degeneration in osteoarthritis. Faseb J. 2022;36(6):e22377. [19] MEINI S, CUCCHI P, CATALANI C, et al. Bradykinin and B₂ receptor antagonism in rat and human articular chondrocytes. Br J Pharmacol. 2011;162(3):611-622. [20] ARELLANO H, NARDELLO-RATAJ V, SZUNERITS S, et al. Saturated long chain fatty acids as possible natural alternative antibacterial agents: Opportunities and challenges. Adv Colloid Interface Sci. 2023;318:102952. [21] RIEGGER J, SCHOPPA A, RUTHS L, et al. Oxidative stress as a key modulator of cell fate decision in osteoarthritis and osteoporosis: a narrative review. Cell Mol Biol Lett. 2023;28(1):76. [22] HUANG Z, KRAUS VB. Does lipopolysaccharide-mediated inflammation have a role in OA?. Nat Rev Rheumatol. 2016;12(2):123-129. [23] AN X, WANG T, ZHANG W, et al. Chondroprotective Effects of Combination Therapy of Acupotomy and Human Adipose Mesenchymal Stem Cells in Knee Osteoarthritis Rabbits via the GSK3β-Cyclin D1-CDK4/CDK6 Signaling Pathway. Aging Dis. 2020;11(5):1116-1132. [24] BHOGE SS, SAMAL S. Physiotherapeutic Approach for Septic Arthritis of Knee Joint. Cureus. 2023;15(9):e45550. [25] 冯方,刘英,彭毅,等.膝骨关节炎患者关节滑液中IL-17水平与疾病严重程度的相关性[J].西部医学,2020,32(6):845-848. [26] IIJIMA H, GILMER G, WANG K, et al. Meta-analysis Integrated With Multi-omics Data Analysis to Elucidate Pathogenic Mechanisms of Age-Related Knee Osteoarthritis in Mice. J Gerontol A Biol Sci Med Sci. 2022;77(7):1321-1334. [27] 徐以明,薛松,桑伟林,等.巨噬细胞极化在骨关节炎中的作用[J].中国骨与关节杂志,2020,9(9):689-694. [28] BARBOZA E, HUDSON J, CHANG WP, et al. Profibrotic Infrapatellar Fat Pad Remodeling Without M1 Macrophage Polarization Precedes Knee Osteoarthritis in Mice With Diet-Induced Obesity. Arthritis Rheumatol. 2017;69(6):1221-1232. [29] HAUBRUCK P, PINTO MM, MORADI B, et al. Monocytes, Macrophages, and Their Potential Niches in Synovial Joints - Therapeutic Targets in Post-Traumatic Osteoarthritis? Front Immunol. 2021;12: 763702. [30] ROMERA-CáRDENAS G, THOMAS LM, LOPEZ-COBO S, et al. Ionomycin Treatment Renders NK Cells Hyporesponsive. PLoS One. 2016;11(3):e0150998. [31] SOHN HS, CHOI JW, JHUN J, et al. Tolerogenic nanoparticles induce type II collagen-specific regulatory T cells and ameliorate osteoarthritis. Sci Adv. 2022; 8(47):eabo5284. [32] HSIEH JL, SHIAU AL, LEE CH, et al. CD8+ T cell-induced expression of tissue inhibitor of metalloproteinses-1 exacerbated osteoarthritis. Int J Mol Sci. 2013;14(10):19951-19970. [33] BECKER J, SEMLER O, GILISSEN C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88(3):362-371. [34] MALEK N, MLOST J, KOSTRZEWA M, et al. Description of Novel Molecular Factors in Lumbar DRGs and Spinal Cord Factors Underlying Development of Neuropathic Pain Component in the Animal Model of Osteoarthritis. Mol Neurobiol. 2024; 61(3):1580-1592. [35] JONES AG, AQUILINO M, TINKER RJ, et al. Clustered de novo start-loss variants in GLUL result in a developmental and epileptic encephalopathy via stabilization of glutamine synthetase. Am J Hum Genet. 2024;111(4):729-741. [36] YANG LG, MARCH ZM, STEPHENSON RA, et al. Apolipoprotein E in lipid metabolism and neurodegenerative disease. Trends Endocrinol Metab. 2023;34(8):430-445. [37] RAULIN C, DOSS SV, TROTTIER ZA, et al. ApoE in Alzheimer’s disease: pathophysiology and therapeutic strategies. Mol Neurodegener. 2022;17(1):72. |
| [1] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [2] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [3] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [4] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [5] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [6] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [7] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [8] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [9] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| [10] | Tian Yushi, Fu Qiang, Li Ji . Bioinformatics identification and validation of mitochondrial genes related to acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6697-6707. |
| [11] | Xie Yizi, Lin Xueying, Zhang Xinxin, Huang Xiufang, Zhan Shaofeng, Jiang Yong, Cai Yan. Biological mechanism of mitophagy in idiopathic pulmonary fibrosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6708-6716. |
| [12] | Cui Yuena, Chen Xiaoyu, Liang Meiting, Chen Wujin, He Yi, Dilinur·Ekpa, Du Manxi, Zhu Yuqiu, Abuduwupuer·Haibier, Sun Yuping. Differences of calorie restriction and time-restricted feeding on metabolic indices and gut microbiota of mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6449-6456. |
| [13] | Zhu Xuekun, Liu Heng, Feng Hui, Gao Yunlong, Wen Lei, Cai Xiaosong, Zhao Ben, Zhong Min. Identification of core genes of osteoarthritis by bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(3): 637-644. |
| [14] | Qiu Boyuan, Liu Fei, Tong Siwen, Ou Zhixue, Wang Weiwei. Bioinformatics identification and validation of aging key genes in hormonal osteonecrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(26): 5608-5620. |
| [15] | Hao Maochen, Ma Chao, Liu Kai, Liu Kexin, Meng Lingting, Wang Xingru, Wang Jianzhong. Bioinformatics screening of key genes for endoplasmic reticulum stress in osteoarthritis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(26): 5632-5641. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||