Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (26): 5608-5620.doi: 10.12307/2025.787
Previous Articles Next Articles
Bioinformatics identification and validation of aging key genes in hormonal osteonecrosis of the femoral head
Qiu Boyuan1, 2, Liu Fei1, Tong Siwen1, Ou Zhixue2, Wang Weiwei1
- 1Graduate School of Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China; 2Department of Joint and Sports Medicine, Guilin Traditional Chinese Medicine Hospital, Guilin 541000, Guangxi Zhuang Autonomous Region, China
-
Received:2024-10-11Accepted:2024-11-12Online:2025-09-18Published:2025-02-25 -
Contact:Ou Zhixue, Chief physician, MD, Professor, Department of Joint and Sports Medicine, Guilin Traditional Chinese Medicine Hospital, Guilin 541000, Guangxi Zhuang Autonomous Region, China Co-corresponding author: Wang Weiwei, Physician, PhD candidate, Graduate School of Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China -
About author:Qiu Boyuan, Master candidate, Graduate School of Guangxi University of Chinese Medicine, Nanning 530000, Guangxi Zhuang Autonomous Region, China; Department of Joint and Sports Medicine, Guilin Traditional Chinese Medicine Hospital, Guilin 541000, Guangxi Zhuang Autonomous Region, China -
Supported by:Guangxi University of Chinese Medicine Graduate Education Innovation Program, No. YCSY2023067 (to QBY); Guangxi Natural Science Foundation Project, No. 2023GXNSFAA026412 (to OZX); Guangxi Zhuang Autonomous Region Traditional Chinese Medicine Administration Self-Financing Scientific Research Project, No. GXZYZ20210372 (to OZX)
CLC Number:
Cite this article
Qiu Boyuan, Liu Fei, Tong Siwen, Ou Zhixue, Wang Weiwei. Bioinformatics identification and validation of aging key genes in hormonal osteonecrosis of the femoral head[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(26): 5608-5620.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

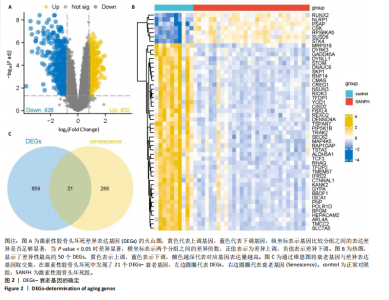
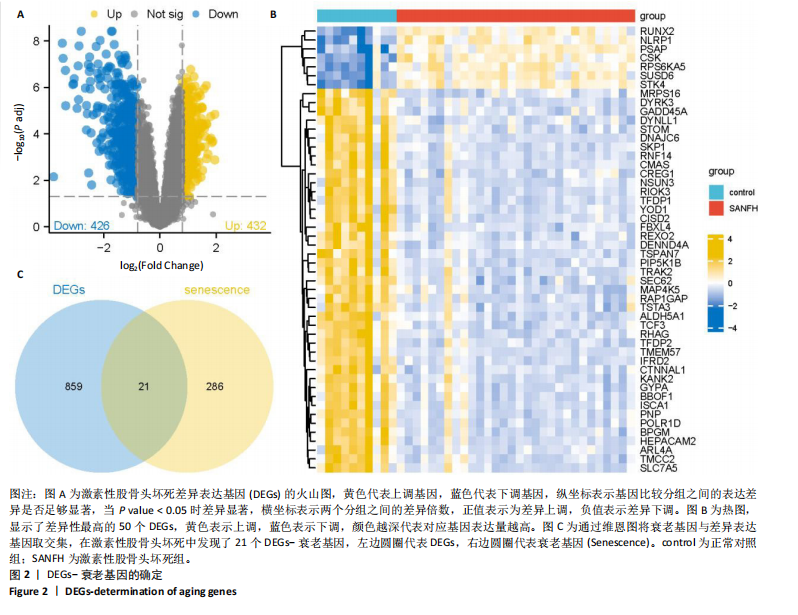
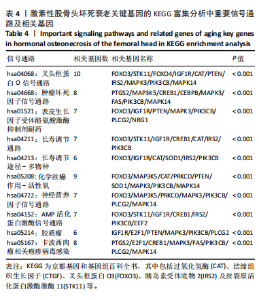
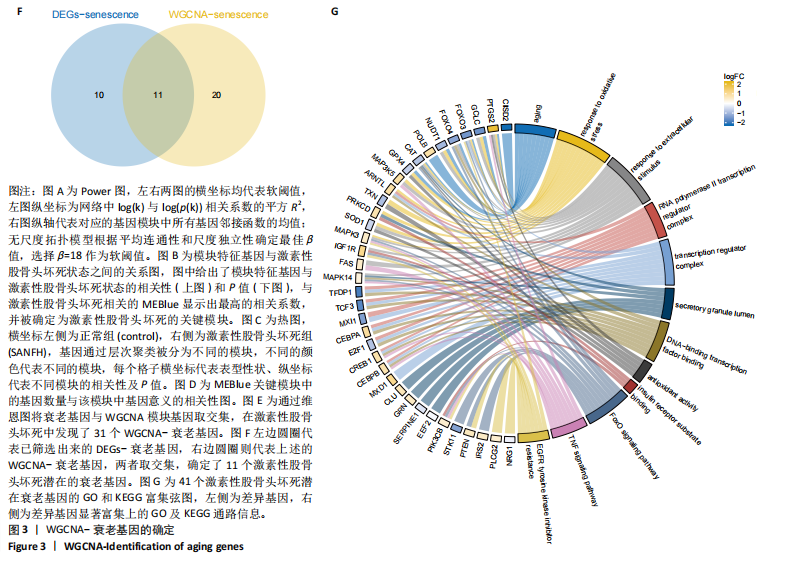
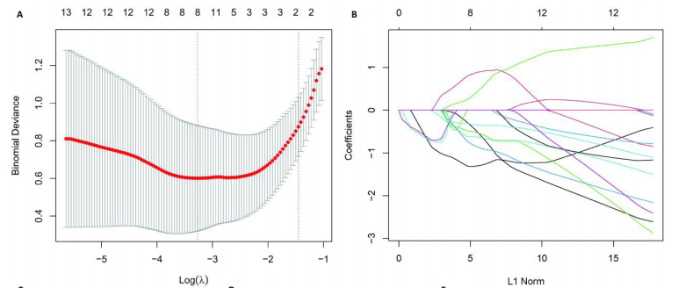
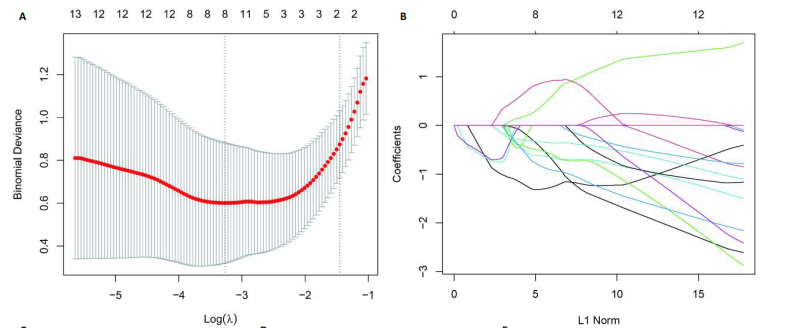
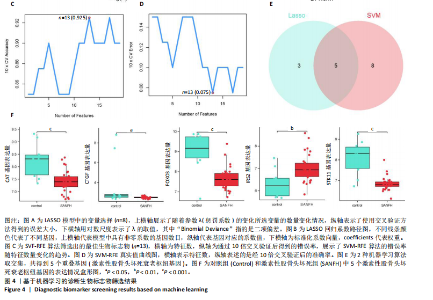
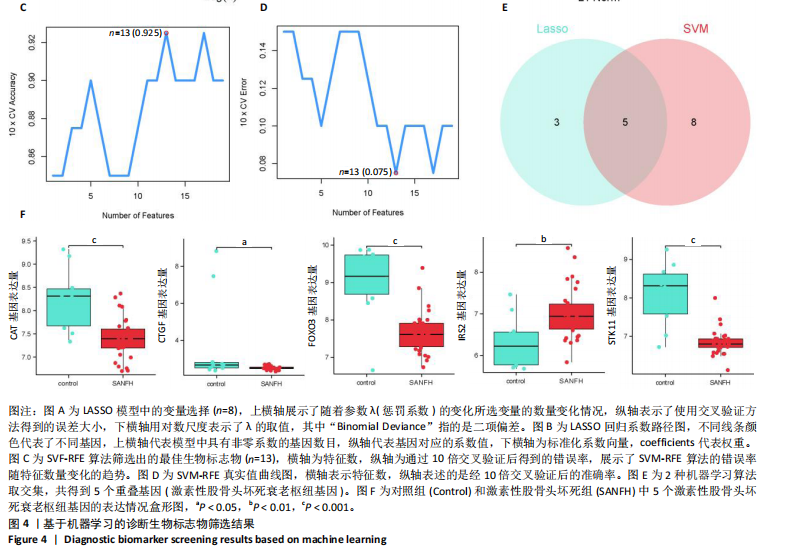
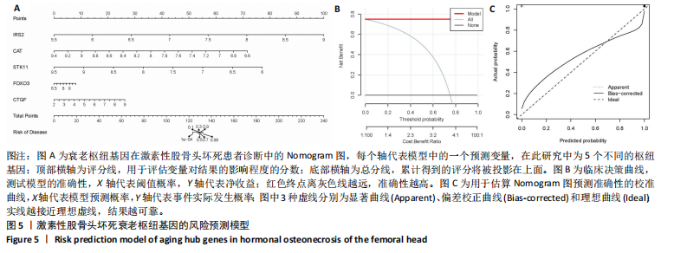
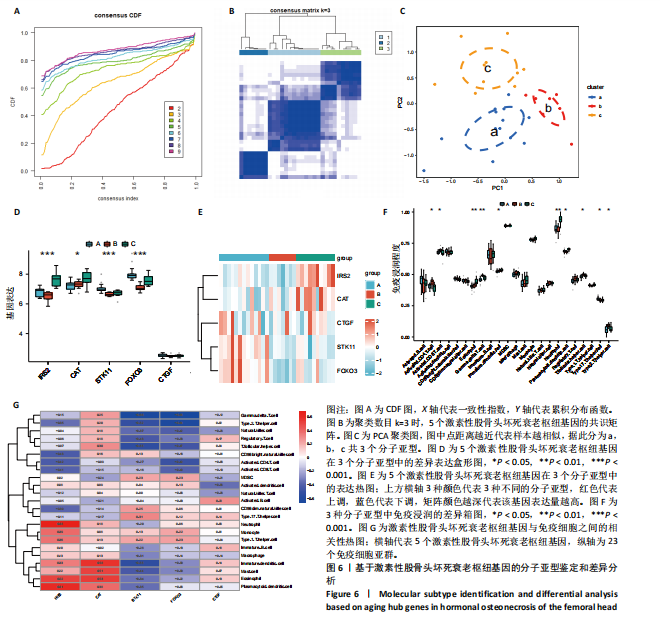
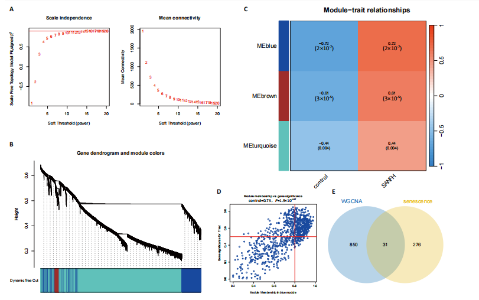
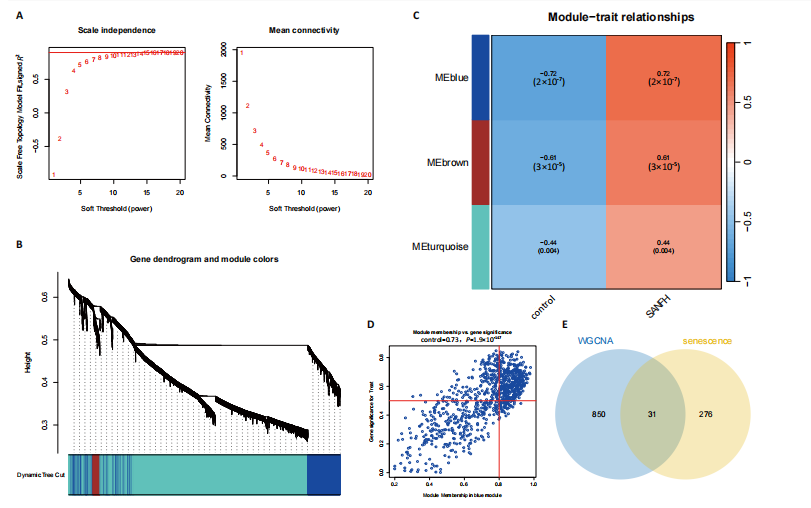
2.1 激素性股骨头坏死衰老基因的差异分析 使用 “limma “R软件包和|logFC|> 0.8和Padj < 0.05的筛选标准鉴定了858个DEGs,其中432个基因在激素性股骨头坏死中上调,426个基因下调。使用热图和火山图描述了差异特征,见图2A,B。图2C显示了激素性股骨头坏死差异基因与衰老相关基因集之间的交集情况。这些交集产生了21个DEG-衰老基因。 2.2 WGCNA分析和激素性股骨头坏死潜在衰老基因的鉴定 使用“WGCNA”R包将前25%基因的表达方差作为筛选条件,剔除波动性较低的基因,并使用4 708个基因构建了共表达网络。选取18作为最佳软阈值(R2=0.9)来构建无标度网络,见图3A。随后,利用聚类分析确定高度相似的模块,最小模块大小设为100个基因。利用动态杂交剪切获得了3个基因模块,其中一个蓝色模块(881个基因)与激素性股骨头坏死的相关性(cor)最高(cor=0.72;P < 0.001),见图3B。此外,在蓝色模块中,GS和MM之间也有很强的相关性(cor=0.73;P < 0.001),见图3D。将蓝色模块中的881个基因与衰老相关基因集取交集,得到了31个WGCNA-衰老基因,见图3E。将DEGs-衰老基因和WGCNA-衰老基因进行取并集,发现了41个激素性股骨头坏死潜在衰老基因,见图3F。GO和KEGG富集分析弦图见图3G,结果表明激素性股骨头坏死潜在衰老基因主要富集在衰老、氧化应激反应、RNA聚合酶Ⅱ转录调节复合物、转录调节复合物、DNA结合转录因子结合、抗氧化活性、FOXO信号通路及肿瘤坏死因子信号通路等方面。具体见表3,4。 2.3 激素性股骨头坏死衰老枢纽基因的鉴定结果 为了进一步明确激素性股骨头坏死衰老枢纽基因,此研究使用了LASSO和SVM-RFE两种机器学习算法来进行鉴定,见图4A-D。结合两种算法的结果(交集),共得到了5个激素性股骨头坏死衰老枢纽基因,它们分别是过氧化氢酶、结缔组织生长因子、叉头框蛋白O3、胰岛素受体底物2和丝裂原活化蛋白激酶激酶11,见图4E。其中,Student’s t检验结果显示,过氧化氢酶、结缔组织生长因子、叉头框蛋白O3 和丝裂原活化蛋白激酶激酶11 在激素性股骨头坏死中低表达,而胰岛素受体底物2在激素性股骨头坏死中高表达,见图4F。 2.4 激素性股骨头坏死衰老枢纽基因风险预测模型的构建 此研究根据激素性股骨头坏死衰老枢纽基因的表达建立了一个激素性股骨头坏死诊断nomogram图,以获得一个更适用于临床的激素性股骨头坏死诊断模型,见图5A。通过构建该模型的临床决策曲线和临床校准曲线,明显可见该模型对激素性股骨头坏死具有较高的预测能力,见图5B,C。 2.5 不同分子亚型之间的差异分析和免疫浸润分析结果 累积分布函数(Cumulative Distribution Function,CDF)图显示当k=3时波动较小,共识矩阵的热图显示了清晰明确的边界,见图6A,B。PCA图显示,a,b和c 组显著区分"

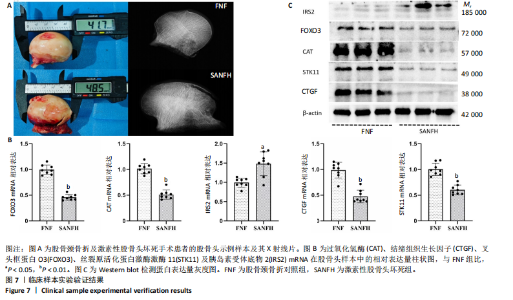
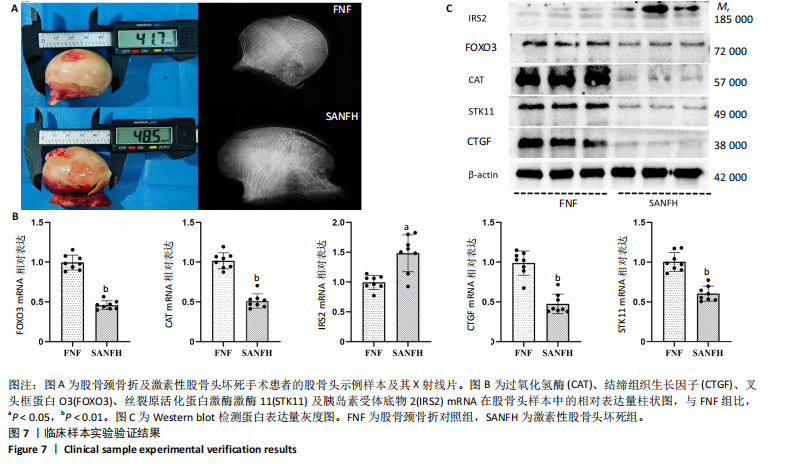
细胞、Th17细胞及Th2细胞的丰度在3个分子亚类中有显著差异,见图 6F。图6G显示了5个激素性股骨头坏死衰老中枢基因与 23个免疫细胞之间的相关性。 2.6 枢纽基因的实验验证结果 通过生物信息数据分析明确了激素性股骨头坏死衰老枢纽基因(过氧化氢酶、结缔组织生长因子、叉头框蛋白O3、胰岛素受体底物2和丝裂原活化蛋白激酶激酶11),并基于Nomogram模型,验证了衰老枢纽基因对激素性股骨头坏死诊断的预测能力。为了进一步验证上述结果,使用股骨头临床样本进行实验验证。其中股骨颈骨折需进行人工全髋关节置换患者的股骨头作为对照组,激素性股骨头坏死需进行人工全髋关节置换患者的股骨头作为激素性股骨头坏死组,见图7A。 对两组共16个样本提取RNA进行qPCR检测,每组分别随机挑选3个样本并提取蛋白进行Western blot验证。qPCR的结果显示,与对照组相比衰老枢纽基因过氧化氢酶(P < 0.01)、结缔组织生长因子(P < 0.01)、叉头框蛋白O3(P < 0.01)和丝裂原活化蛋白激酶激酶11(P < 0.01)"
| [1] ZHANG J, CAO J, LIU Y, et al. Advances in the pathogenesis of steroid-associated osteonecrosis of the femoral head. Biomolecules. 2024; 14(6):667. [2] LIJC, LIANG XZ, LUO D, et al. Study on the molecular mechanism of BuShenHuoXue capsule in treatment of steroid-induced osteonecrosis ofthefemoral head. Ann Transl Med. 2020;8(24):1680. [3] LUCAS V, CAVADAS C, AVELEIRA CA. Cellular senescence: from mechanisms to current biomarkers and senotherapies. Pharmacol Rev. 2023;75(4):675-713. [4] LI X, LI C, ZHANG W, et al. Inflammation and aging: signaling pathwaysand intervention therapies. Signal Transduct Target Ther. 2023;8(1):239. [5] FULOP T, LARBI A, PAWELEC G, et al. Immunology of aging: the birth of inflammaging. Clin Rev Allergy Immunol. 2023;64(2):109-122. [6] MOERMAN EJ, TENG K, LIPSCHITZ DA, et al. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;(6):3. [7] CHEN H, LIU X, CHEN H, et al. Role of SIRT1 and AMPK in mesenchymal stem cells differentiation. Ageing Res Rev. 2014;(13):55-64. [8] TANG B, CHEN Y, ZHAO P, et al. MiR-601-induced BMSCs senescence accelerates steroid-induced osteonecrosis of the femoral head progression by targeting SIRT1. Cell Mol Life Sci. 2023;80(9):261. [9] TRAN KA, KONDRASHOVA O, BRADLEY A, et al. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13(1):152. [10] WALJEE AK, WEINHEIMER-HAUS EM, ABUBAKAR A, et al. Artificial intelligence and machine learning for early detection and diagnosis of colorectal cancer in sub-Saharan Africa. Gut. 2022;71(7):1259-1265. [11] GREENER JG, KANDATHIL SM, MOFFAT L, et al. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23(1):40-55. [12] ZHANG F, PENG WX, WANG L, et al. Role of FGF-2 transfected bone marrow mesenchymal stem cells in engineered bone tissue for repair of avascular necrosis of femoral head in rabbits. Cell Physiol Biochem. 2018;48(2):773-784. [13] AVELAR RA, ORTEGA JG, TACUTU R, et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21(1):91. [14] LI Y, LU S, LAN M, et al. A prognostic nomogram integrating novel biomarkers identified by machine learning for cervical squamous cell carcinoma. J Transl Med. 2020;18(1):223. [15] LI G, LI X, YANG M, et al. Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci Rep. 2017;7:42105. [16] KANEHISA M, FURUMICHI M, SATO Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545-D551. [17] SIMON N, FRIEDMAN J, HASTIE T, et al. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011; 39(5):1-13. [18] CHEN D, LIU J, ZANG L, et al. Integrated machine learning and bioinformatic analyses constructed a novel stemness-related classifier to predict prognosis and immunotherapy responses for hepatocellular carcinoma patients. Int J Biol Sci. 2022;18(1):360-373. [19] CHEN T, ZHANG H, LIU Y, et al. EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J Genet Genomics. 2021;48(9):863-866. [20] WILKERSON MD, HAYES DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572-1573. [21] BARBIE DA, TAMAYO P, BOEHM JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108-112. [22] SUO J, SHAO R, YANG R, et al. Accelerated aging in articular cartilage by ZMPSTE24 deficiency leads to osteoarthritis with impaired metabolic signaling and epigenetic regulation. Cell Death Dis. 2023;14(5):336. [23] OKAMOTO M, NAKASHIMA H, SAKAI K, et al. Cellular senescence is associated with osteonecrosis of the femoral head while mesenchymal stem cell conditioned medium inhibits bone collapse. Sci Rep. 2024; 14(1):3329. [24] CHEN X, GONG W, SHAO X, et al. METTL3-mediated m6A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022;81(1):87-99. [25] BAKER A, LIN CC, LETT C, et al. Catalase: a critical node in the regulation of cell fate. Free Radic Biol Med. 2023;199:56-66. [26] ABDALBAGEMOHAMMEDABDALSADEG S, XIAO BL, MA XX, et al. Catalase immobilization: current knowledge, key insights, applications, and future prospects - A review. Int J Biol Macromol. 2024;276(Pt 2): 133941. [27] NDREPEPA G. Myeloperoxidase – A bridge linking inflammation and oxidative stress with cardiovascular disease. Clinica Chimica Acta. 2019;493:36-51. [28] SALMINEN A.Immunosuppressive network promotes immunosenescence associated withaging and chronic inflammatory conditions. J Mol Med (Berl). 2021:99(11):1553-1569. [29] HU K, SHANG Z, YANG X, et al. Macrophage Polarization and the Regulation of Bone Immunity in Bone Homeostasis. J Inflamm Res. 2023;16:3563-3580. [30] CHIANG CC, CHENG WJ, KORINEK M, et al. Neutrophils in psoriasis. Front Immunol. 2019;10:2376. [31] COWLAND JB, BORREGAARD N. Granulopoiesis and granules of human neutrophils. Immunol Rev. 2016;273(1):11-28. [32] CALNAN DR, BRUNET A. The FoxO code. Oncogene. 2008;27(16): 2276-2288. [33] OMOROU M, HUANG Y, GAO M, et al. The forkhead box O3 (FOXO3): a key player in the regulation of ischemia and reperfusion injury. Cell Mol Life Sci. 2023;80(4):102. [34] CAO G, LIN M, GU W, et al. The rules and regulatory mechanisms of FOXO3 on inflammation, metabolism, cell death and aging in hosts. Life Sci. 2023;328:121877. [35] ATASHI F, MODARRESSI A, PEPPER MS. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cells Dev. 2015;24(10):1150-1163. [36] ADAM-VIZI V, CHINOPOULOS C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci. 2006; 27(12):639-645. [37] GOMEZ-PUERTO M, VERHAGEN LP, BRAAT AK, et al. Activation of autophagy by FOXO3 regulates redox homeostasis during osteogenic differentiation. Autophagy. 2016;12(10):1804-1816. [38] JEONG SG, CHO GW. Endogenous ROS levels are increased in replicative senescence in human bone marrow mesenchymal stromal cells. Biochem Biophys Res Commun. 2015;460(4):971-976. [39] ZHOU T, YAN Y, ZHAO C, et al. Resveratrol improves osteogenic differentiation of senescent bone mesenchymal stem cells through inhibiting endogenous reactive oxygen species production via AMPK activation. Redox Rep. 2019;24(1):62-69. [40] KANAZAWA I. Interaction between bone and glucose metabolism. Endocr J. 2017;64(11):1043-1053. [41] LIAN WS, WU RW, CHEN YS, et al. MicroRNA-29a mitigates osteoblast senescence and counteracts bone loss through oxidation resistance-1 control of FoxO3 methylation. Antioxidants (Basel). 2021;10(8):1248. [42] XI G, WAI C, WHITE MF, et al. Down-regulation of insulin receptor substrate 1 during hyperglycemia induces vascular smooth muscle cell dedifferentiation. J Biol Chem. 2017;292(5):2009-2020. [43] ZHAO H, LIU F, JIA R, et al. MiR-570 inhibits cell proliferation and glucose metabolism by targeting IRS1 and IRS2 in human chronic myelogenous leukemia. Iran J Basic Med Sci. 2017;20(5):481-488. [44] ZHANG H, WANG Z, LI Q, et al. IRTKS promotes osteogenic differentiation by inhibiting PTEN phosphorylation. Biomed Pharmacother. 2024;177:116872. [45] LI B, WANG Y, LIU Y, et al. Altered gene expression involved in insulin signaling pathway in type II diabetic osteoporosis rats model. Endocrine. 2013;43(1):136-146. [46] CLAASSEN H, SCHLÜTER M, SCHÜNKE M, et al. Influence of 17beta-estradiol and insulin on type II collagen and protein synthesis of articular chondrocytes. Bone. 2006;39(2):310-317. [47] KELLNER K, SCHULZ MB, GÖPFERICH A, et al. Insulin in tissue engineering of cartilage: a potential model system for growth factor application. J Drug Target. 2001;9(6):439-448. [48] CAI L, OKUMU FW, CLELAND JL, et al. A slow release formulation of insulin as a treatment for osteoarthritis. Osteoarthritis Cartilage. 2002;10(9):692-706. [49] SHAO J, LIU S, ZHENG X, et al. Berberine promotes peri-implant osteogenesis in diabetic rats by ROS-mediated IRS-1 pathway. Biofactors. 2021;47(1):80-92. [50] MIAO M, ZHANG Y, WANG X, et al. The miRNA-144-5p/IRS1/AKT axis regulates the migration, proliferation, and mineralization of osteoblasts: a mechanism of bone repair in diabetic osteoporosis. Cell Biol Int. 2022;46(12):2220-2231. [51] SANO T, OCHIAI T, NAGAYAMA T, et al. Genetic reduction of insulin signaling mitigates amyloid-β deposition by promoting expression of extracellular matrix proteins in the brain. J Neurosci. 2023;43(43): 7226-7241. [52] OCHIAI T, SANO T, NAGAYAMA T, et al. Differential involvement of insulin receptor substrate (IRS)-1 and IRS-2 in brain insulin signaling is associated with the effects on amyloid pathology in a mouse model of Alzheimer’s disease. Neurobiol Dis. 2021;159:105510. [53] REN M, YAO S, CHEN T, et al. Connective tissue growth factor: regulation, diseases, and drug discovery. Int J Mol Sci. 2024;25(9):4692. [54] SHEN YW, ZHOU YD, CHEN HZ, et al. Targeting CTGF in cancer: an emerging therapeutic opportunity. Trends Cancer. 2021;7(6):511-524. [55] YU L, WEN H, LIU C, et al. Embryonic stem cell-derived extracellular vesicles rejuvenate senescent cells and antagonize aging in mice. Bioact Mater. 2023;29:85-97. [56] NISHIDA T, EMURA K, KUBOTA S, et al. CCN family 2/connective tissue growth factor (CCN2/CTGF) promotes osteoclastogenesis via induction of and interaction with dendritic cell-specific transmembrane protein (DC-STAMP). J Bone Miner Res. 2011;26(2):351-363. [57] PONS-TOSTIVINT E, LUGAT A, FONTENAU JF, et al. STK11/LKB1 Modulation of the immune response in lung cancer: from biology to therapeutic impact. Cells. 2021;10(11):3129. [58] TRELFORD CB, SHEPHERD TG. LKB1 biology: assessing the therapeutic relevancy of LKB1 inhibitors. Cell Commun Signal. 2024;22(1):310. [59] LI J, WANG Y, CHEN D, et al. Oral administration of berberine limits post-traumatic osteoarthritis development and associated pain via AMP-activated protein kinase (AMPK) in mice. Osteoarthritis Cartilage. 2022;30(1):160-171. [60] ZHENG Y, TAO Y, ZHAN X, et al. Nuclear receptor 4A1 (NR4A1) silencing protects hepatocyte against hypoxia-reperfusion injury in vitro by activating liver kinase B1 (LKB1)/AMP-activated protein kinase (AMPK) signaling. Bioengineered. 2022;13(4):8349-8359. [61] WANG P, MA T, GUO D, et al. Metformin induces osteoblastic differentiation of human induced pluripotent stem cell-derived mesenchymal stem cells. J Tissue Eng Regen Med. 2018;12(2):437-446. [62] CHEN C, ZHAO X, LUO Y, et al. Imbalanced t-cell subsets may facilitate the occurrence of osteonecrosis of the femoral head. J Inflamm Res. 2022;15:4159-4169. [63] ELSAYED R, KURAGO Z, CUTLER CW, et al. Role of dendritic cell-mediated immune response in oral homeostasis: a new mechanism of osteonecrosis of the jaw. FASEB J. 2020;34(2):2595-2608. [64] WANG B, DONG Y, TIAN Z, et al. The role of dendritic cells derived osteoclasts in bone destruction diseases. Genes Dis. 2020;8(4): 401-411. [65] OGAWA H, YOKATA S, HOSOI Y, et al. Methylprednisolone pulse-enhanced neutrophil extracellular trap formation in mice with imiquimod-induced lupus-like disease, resulting in ischaemia of the femoral head cartilage. Lupus Sci Med. 2023;10(2):e001042. |
| [1] | Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. The construction and validation of a prediction model based on multiple machine learning algorithms and the immunomodulatory analysis of rheumatoid arthritis from the perspective of mitophagy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-15. |
| [2] | Miao Jiahang, Ma Sheng, Li Qupeng, Yu Huilin, Hu Tianyu, Gao Xiao, Feng Hu. Cervical lordosis ratio can be used as a decision-making indicator for selection of posterior surgical approach for multi-level cervical spondylotic myelopathy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1796-1802. |
| [3] | Lyu Liting, Yu Xia, Zhang Jinmei, Gao Qiaojing, Liu Renfan, Li Meng, Wang Lu. Bibliometric analysis of research process and current situation of brain aging and exosomes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1457-1465. |
| [4] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [5] | Chen Yilin, Jiang Xiaobo, Qu Honglin, Liu Ruilian. General pattern of GSK3/Nrf2-regulated biological rhythms in organismal aging [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1257-1264. |
| [6] | Wen Zixing, Xu Xin, Zhu Shengqun. Correlations between gastrocnemius morphology parameters and physical activity capacity in elderly females under high-frequency ultrasound [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1058-1063. |
| [7] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [8] | Yang Bin, Tao Guangyi, Yang Shun, Xu Junjie, Huang Junqing . Visualization analysis of research hotspots of artificial intelligence in field of spinal cord nerve injury and repair [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(4): 761-770. |
| [9] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| [10] | Zhang Xiongjinfu, Chen Yida, Cheng Xinyi, Liu Daihui, Shi Qin . Exosomes derived from bone marrow mesenchymal stem cells of young rats to reverse senescence in aged rat bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7709-7718. |
| [11] | Sima Xinli, Liu Danping, Qi Hui. Effect and mechanism of metformin-modified bone marrow mesenchymal stem cell exosomes on regulating chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7728-7734. |
| [12] | Li Jing, Lu Guangqi, Zhuang Minghui, Cui Ying, Yu Zhangjingze, Sun Xinyue, Ma Mingming, Zhu Liguo, Yu Jie. Development of a clinical prediction model for cervical instability in young and middle-aged adults based on machine learning [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(33): 7203-7210. |
| [13] | Wang Simin, Zhang Dezhou, Zhao Jing, Wang Chaoqun, Li Kun, Chen Jie, Bai Xue, Zhao Hailong, Zhang Shaojie, Ma Yuan, Hao Yunteng, Yang Yang, Li Zhijun, Shi Jun, Wang Xing. Artificial intelligence and cervical spine image recognition: application prospects and challenges [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(33): 7231-7240. |
| [14] | Tian Yushi, Fu Qiang, Li Ji . Bioinformatics identification and validation of mitochondrial genes related to acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6697-6707. |
| [15] | Xie Yizi, Lin Xueying, Zhang Xinxin, Huang Xiufang, Zhan Shaofeng, Jiang Yong, Cai Yan. Biological mechanism of mitophagy in idiopathic pulmonary fibrosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6708-6716. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||