Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (3): 770-784.doi: 10.12307/2026.009
Previous Articles Next Articles
An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation
Zhao Feifan1, Cao Yujing2
- 1College of Bone Injury, Henan University of Traditional Chinese Medicine, Zhengzhou 450000, Henan Province, China; 2Henan Provincial Hospital of Traditional Chinese Medicine, Zhengzhou 450003, Henan Province, China
-
Received:2024-11-06Accepted:2024-12-14Online:2026-01-28Published:2025-07-10 -
Contact:Cao Yujing, PhD, Professor, Henan Provincial Hospital of Traditional Chinese Medicine, Zhengzhou 450003, Henan Province, China -
About author:Zhao Feifan, Master candidate, College of Bone Injury, Henan University of Traditional Chinese Medicine, Zhengzhou 450000, Henan Province, China -
Supported by:Henan Provincial Administration of Traditional Chinese Medicine Scientific Research Special Projects, Nos. 2024ZYZD06 and 2023ZY1008 (to CYJ)
CLC Number:
Cite this article
Zhao Feifan, Cao Yujing. An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 770-784.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
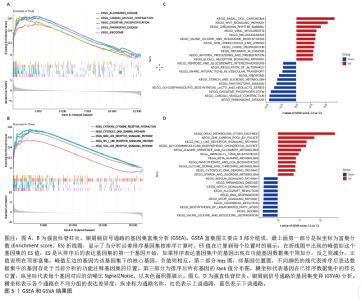
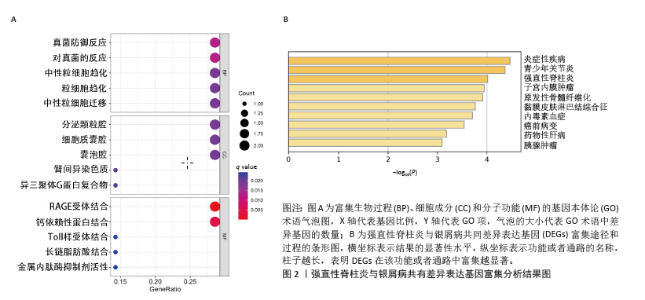
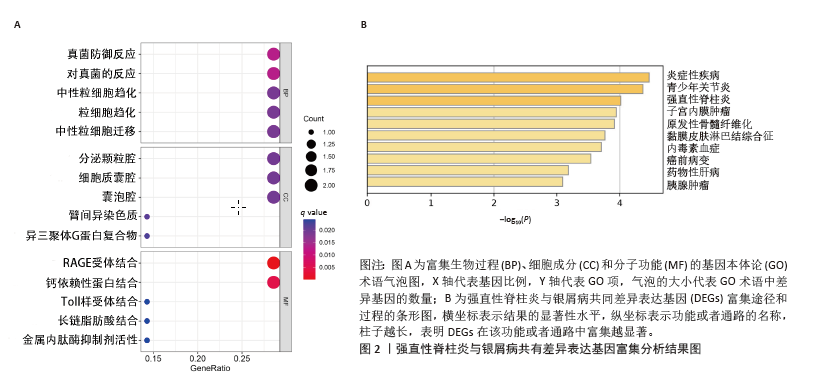
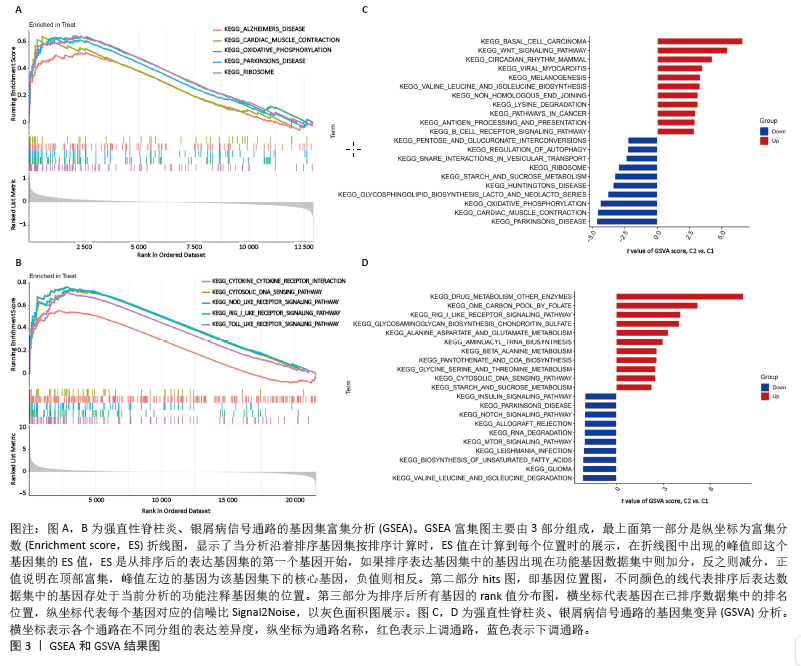
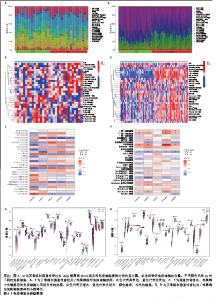
2.3 GSEA和GSVA分析结果 为了进一步阐明强直性脊柱炎/银屑病组和正常组之间的功能差异,进行了GSEA和GSVA分析。GSEA结果发现强直性脊柱炎组主要与氧化磷酸化和核糖体相关的信号通路有关,在KEGG_ALZHEIMERS_DISEASE,KEGG_CARDIAC_MUSCLE_CONTRACTION,KEGG_OXIDATIVE_PHOSPHORYLATION,KEGG_PARKINSONS_DISEASE及KEGG_RIBOSOME通路中显著富集,见图3A;银屑病组主要与细胞DNA和受体信号传导通路有关,在KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION,KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY,KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY,KEGG_RIG_I_LIKE_RECEPTOR_SIGNALING_PATHWAY及KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY通路中显著富集,见图3B;这凸显了细胞器和受体信号通路在强直性脊柱炎/银屑病中的重要参与。结合这些结果,对强直性脊柱炎与银屑病的发病机制进行基因层面的进一步分析十分必要。GSVA结果显示,在强直性脊柱炎中,WNT信号通路、缬氨酸、亮氨酸和异亮氨酸的生物合成、非同源末端连接、赖氨酸降解、抗原处理和呈递、B细胞受体信号通路显著上调,而戊糖和葡萄糖醛酸相互转化、自噬调节、核糖体、淀粉和蔗糖代谢、氧化磷酸化通路则明显下调,见图3C;在银屑病中,叶酸一碳库、RIG-Ⅰ样受体信号通路、硫酸软骨素氨基葡聚糖、丙氨酸、天冬氨酸和谷氨酸代谢、氨酰核糖核酸生物合成、胞质DNA感应通路、淀粉和蔗糖代谢通路显著上调,NOTCH信号通路、RNA降解、MTOR信号通路、不饱和脂肪酸生物合成、缬氨酸和异亮氨酸降解通路显著下调,见图3D。"
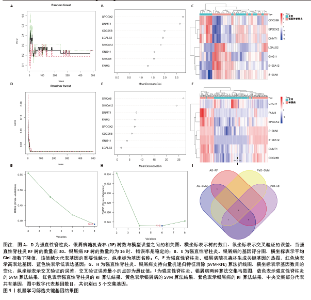
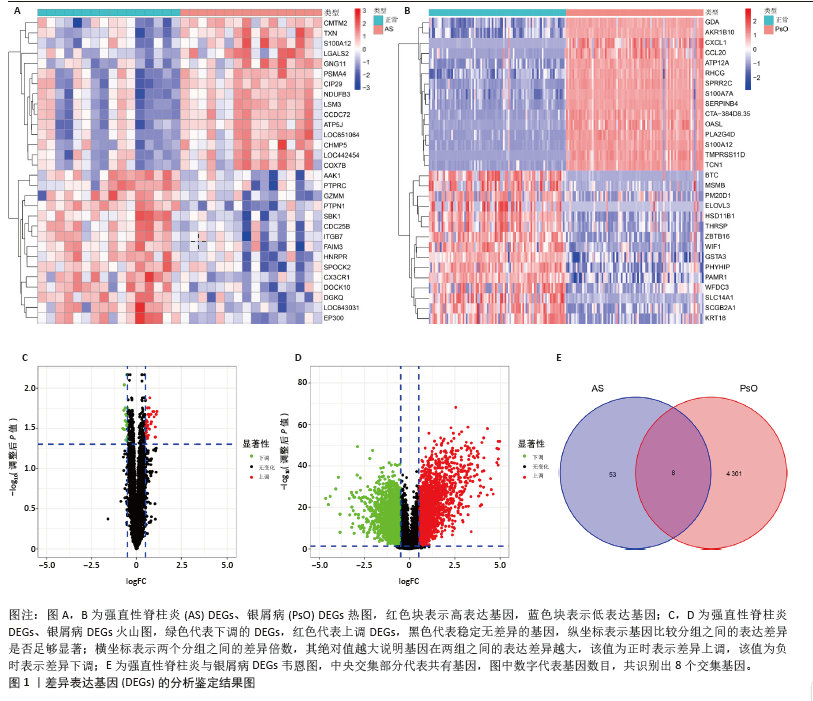
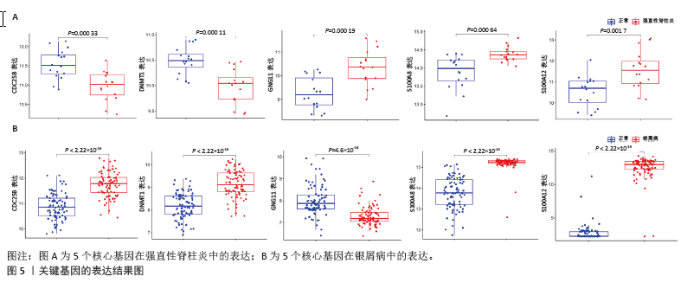
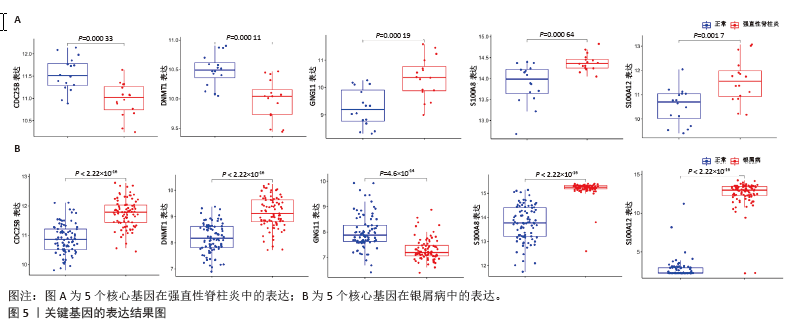
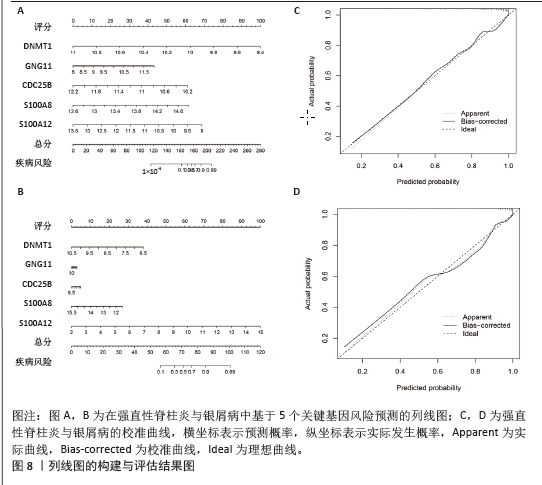
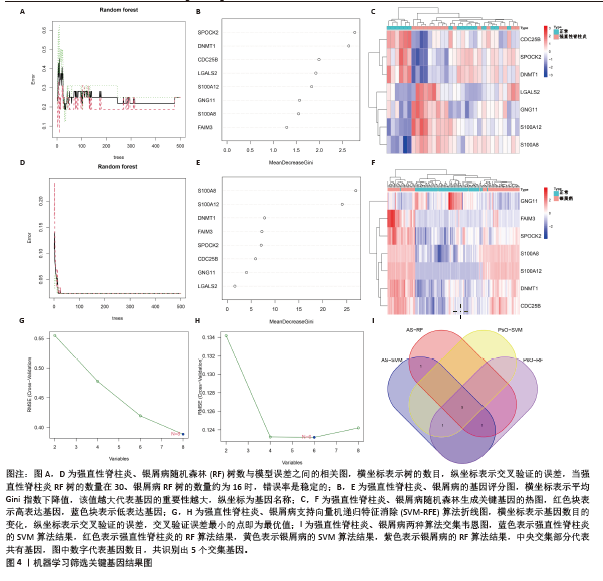
2.4 机器学习筛选关键基因 为了进一步筛选最具诊断价值的关键基因,基于机器学习算法选择了最重要的特征。对上述8个候选基因依次进行SVM-RFE和RF分析。在RF算法中,根据模型误差与决策树数量的关系图,见图4A,D。对于强直性脊柱炎选择了 30 棵决策树作为最终模型的参数,对于银屑病选择了 16 棵决策树作为最终模型的参数,这表明模型误差比较稳定;随后,将强直性脊柱炎中重要度大于 1.5和银屑病中重要度大于 3的 DEGs 作为候选基因进行后续分析,见图4B,E。基于筛选出的重要变量,对 GSE25101 和 GSE30999进行了 k-means 无监督聚类。图4C显示,在强直性脊柱炎数据集的32个样本中,这7个基因可用于区分强直性脊柱炎和正常样本;同样,在银屑病数据集的170个样本中,这7个基因也可用于区分银屑病和正常样本,见图4F。最终通过SVM-RFE筛选出了强直性脊柱炎数据集中的8个基因和银屑病数据集中的6个基因,见图4G,H。RF法筛出了强直性脊柱炎数据集中的7个基因和银屑病数据集中的7个基因。针对上述不同方法在不同数据集中筛选出的基因相互重叠,最终确定了5个共同的生物标志物:DNA甲基转移酶1 (DNMT1)、G蛋白γ11亚基(GNG11)、细胞分裂周期25B (CDC25B)、钙结合蛋白A8 (S100A8)、钙结合蛋白A12(S100A12),见图4I。"
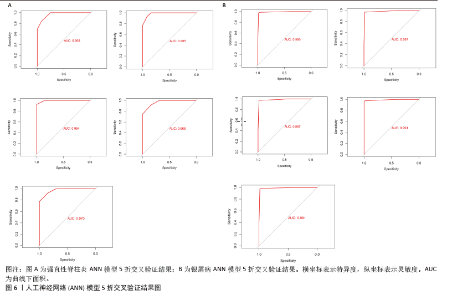
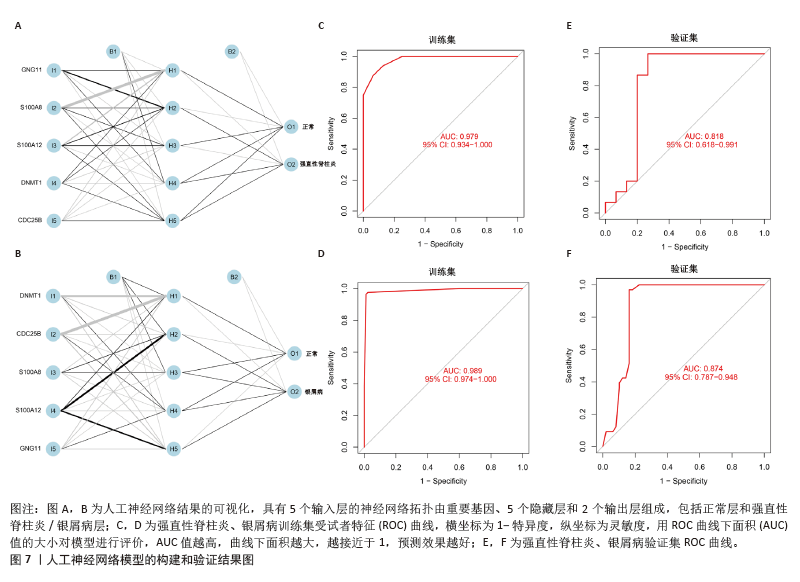
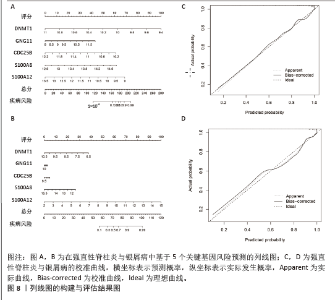
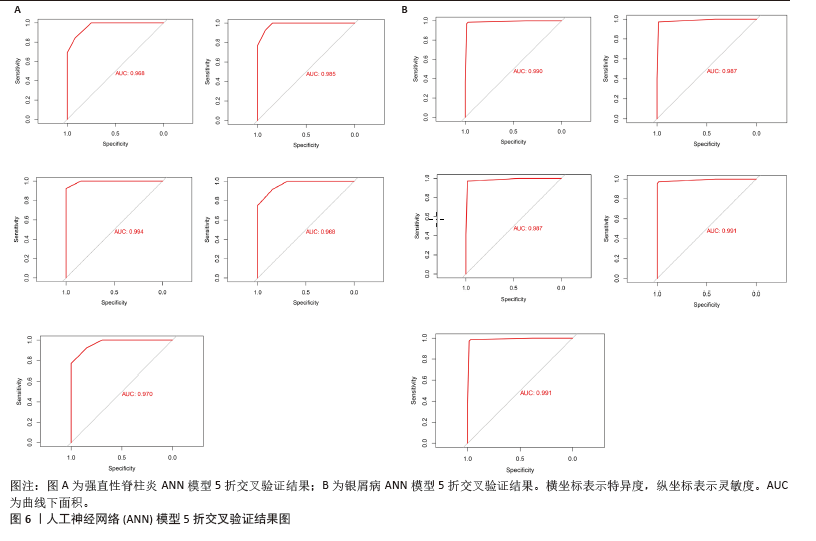
2.6 人工神经网络模型的构建及验证结果 将上述机器学习筛选结果转化为基因评分表,基于基因评分表构建强直性脊柱炎与银屑病的人工神经网络模型。导入5个共有关键基因的评分数据作为输入层,设置5个节点作为中间隐藏层,输出层包括两个节点(疾病/对照)。5折交叉验证的每个结果都用ROC曲线表示,见图6,准确度等指标见表2。5折交叉验证结果的平均AUC超过0.98,证明了该模型的可靠性。该模型在GSE25101和GSE30999中的整体AUC为0.979及0.989,见图7A-D,表明模型性能良好。为了进一步验证人工神经网络模型的准确性,还在外部数据集GSE73754和GSE14905中进行验证。结果显示,在GSE73754中,AUC为0.818,见图7E;在GSE14905中,AUC为0.874,见图7F。"
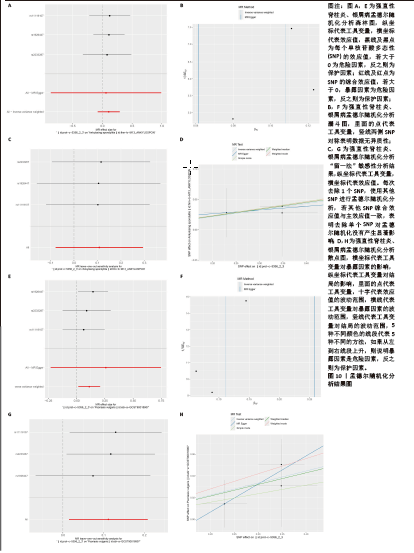
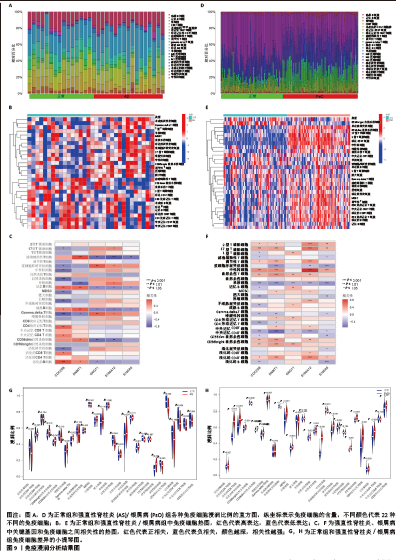
2.8 免疫浸润分析结果 由于富集分析表明免疫对这两种疾病的发展都至关重要,为了探索强直性脊柱炎与银屑病患者的免疫微环境,首先使用CIBERSORT算法估计22种免疫细胞的相对丰度,然后基于免疫相关基因集进行ssGSEA分析,结果见图9A-F。组织免疫细胞浸润分析揭示了强直性脊柱炎数据集32个样本和银屑病数据集170个样本的基因集合中的22种免疫细胞亚型,见图9A,D。图9B,E显示了GSE25101、GSE30999样本中28个免疫细胞的分布。差异表达分析结果显示,与正常样品相比,强直性脊柱炎中活化树突状细胞、CD56bright自然杀伤细胞、γδT细胞、巨噬细胞、调节性T细胞、17型T 辅助细胞浸润更高,而自然杀伤T细胞、滤泡辅助性T细胞、2型T辅助细胞、CD4效应记忆T细胞、中央记忆 CD8 T 细胞浸润显著降低,见图9G;在银屑病中活化CD4T细胞、活化CD8T细胞、活化树突状细胞、CD56dim自然杀伤细胞、嗜酸性粒细胞、未成熟B细胞、MDSC、巨噬细胞、自然杀伤T细胞、自然杀伤细胞、中性粒细胞、浆细胞样树突状细胞、调节性T细胞、滤泡辅助性T细胞、17型T 辅助细胞、2型T 辅助细胞、CD4效应记忆T细胞、中央记忆 CD4 T 细胞浸润显著增高,而活化B细胞、CD56bright自然杀伤细胞、γδT细胞、不成熟树突状细胞、记忆B细胞、CD8效应记忆T细胞浸润显著降低,见图9H。该结果表明强直性脊柱炎与银屑病似乎都发生了免疫失调和炎症反应。通过相关性分析发现,在强直性脊柱炎数据集中,CDC25B与活化B细胞呈显著正相关,与自然杀伤细胞、γδT细胞呈显著负相关;DNMT1与滤泡辅助性T细胞呈显著正相关,与单核细胞呈显著负相关;GNG11与浆细胞样树突状细胞、γδT细胞呈正相关,与滤泡辅助性T细胞、CD56dim自然杀伤细胞呈负相关;S100A12、S100A8与滤泡辅助性T细胞呈负相关,见图9C。在银屑病数据集中,CDC25B与中性粒细胞呈显著正相关,与单核细胞、肥大细胞、CD8效应记忆T细胞、中央记忆 CD8 T 细胞呈负相关;DNMT1与中性粒细胞和自然杀伤细胞呈显著正相关,与中央记忆 CD8 T 细胞呈显著负相关;GNG11与CD56bright自然杀伤细胞呈显著正相关,与单核细胞呈显著负相关;S100A12与中性粒细胞呈显著正相关,与中央记忆 CD8 T 细胞呈负相关;S100A8与中性粒细胞呈显著正相关,与单核细胞、活化B细胞呈负相关,见图9F。该结果表明这些关键基因可能通过调节免疫细胞的表达参与调节自身免疫。 2.9 孟德尔随机化分析结果 为了进一步探究核心基因与强直性脊柱炎与银屑病是否具有因果关系,选择连接节点数目最多的基因S100A8进行后续的孟德尔随机化分析,以期可以更好地识别和理解基因与表型之间的复杂关系,从而增强孟德尔随机化分析的生物学解释力,见图10A-H。在强直性脊柱炎中,森林图显示,rs11119107,rs1926447,rs2233287均位于置信区间右侧,表明存在正相关,随着S100A8表达增加,强直性脊柱炎的发病风险也会增加,见图10A。在银屑病中,rs1926447,rs2233287,rs11119107也均位于置信区间右侧,同样表明存在正相关,即随着S100A8表达增加,银屑病的发病风险会增加,见图10E。进一步剖析固有的异质性,针对强直性脊柱炎与银屑病定制的漏斗图揭示了与预期对称分布的偏差,尽管保持了总体对称性,见图10B,F。通过敏感性分析,采用“留一法”方法进一步审查了这一细致入微的观察结果。值得注意的是,分析中省略任何单个 SNP 对IVW分析结果的影响可以忽略不计,这表明其余 SNP 始终反映了总体数据集的结果,见图10C,G。为了证实研究结果的有效性,调用MR-Egger 回归分析,从而增强结果和所应用的方法的稳健性和真实性。该孟德尔随机化分析明确证实了S100A8和强直性脊柱炎与银屑病的密切关联,见图10D,H。因此,它描绘了一条通过干预 S100A8功能来调节强直性脊柱炎与银屑病的发生、演变和进展的潜在途径,为治疗干预和更深入地了解疾病机制提供了一条有前景的途径。"
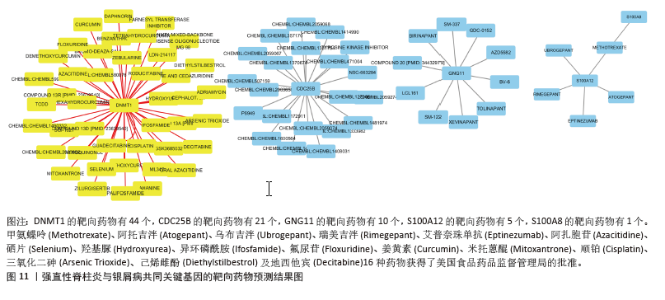
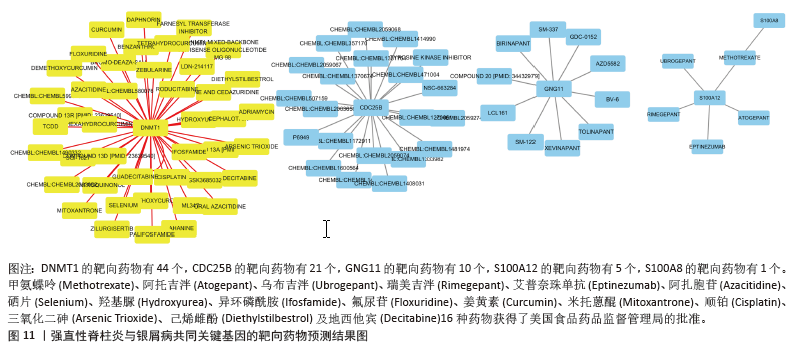
2.10 靶向药物预测结果 DGIdb 共鉴定出81种可能针对强直性脊柱炎与银屑病共同关键基因的分子药物。其中,44个靶向DNMT1、21个靶向CDC25B、10个靶向GNG11、5个靶向S100A12及1个靶向S100A8,见图11。其中,只有甲氨蝶呤(Methotrexate)、阿托吉泮(Atogepant)、乌布吉泮(Ubrogepant)、瑞美吉泮(Rimegepant)、艾普奈珠单抗(Eptinezumab)、阿扎胞苷(Azacitidine)、硒片(Selenium)、羟基脲(Hydroxyurea)、异环磷酰胺(Ifosfamide)、氟尿苷(Floxuridine)、姜黄素(Curcumin)、米托蒽醌(Mitoxantrone)、顺铂(Cisplatin)、三氧化二砷(Arsenic Trioxide)、己烯雌酚(Diethylstilbestrol)及地西他宾(Decitabine)16种药物获得了美国食品药品监督管理局的批准。"
| [1] TAUROG JD, CHHABRA A, COLBERT RA. Ankylosing spondylitis and axial spondyloarthritis. New Engl J Med. 2016; 374(26):2563-2574. [2] WARD MM, DEODHAR A, GENSLER LS, et al. 2019 update of the american college of rheumatology/spondylitis association of america/spondyloarthritis research and treatment network recommendations for the treatment of ankylosing spondylitis and nonradiographic axial spondyloarthritis. Arth Rheum. 2019;71(10):1285-1299. [3] RANGANATHAN V, GRACEY E, BROWN MA, et al. Pathogenesis of ankylosing spondylitis—recent advances and future directions. Nat Rev Rheumatol. 2017;13(6):359-367. [4] CHEN Y, WU Y, YANG H, et al. DNA Methylation and mRNA expression of B7-H3 gene in ankylosing spondylitis: a case-control study. Immunol Invest. 2022;51(7):2025-2034. [5] CHOUDHARY S, KHAN NS, VERMA R, et al. Exploring the molecular underpinning of psoriasis and its associated comorbidities through network approach: cross talks of genes and pathways. 3 Biotech. 2023; 13(5):130. [6] WANG T, XIA Y, ZHANG X, et al. Short-term effects of air pollutants on outpatients with psoriasis in a Chinese city with a subtropical monsoon climate. Front Public Health. 2022; 10:1071263. [7] GUO J, ZHANG H, LIN W, et al. Signaling pathways and targeted therapies for psoriasis. Signal Transduct Target Ther. 2023;8(1):437. [8] GRIFFITHS CEM, ARMSTRONG AW, GUDJONSSON JE, et al. Psoriasis. Lancet. 2021; 397(10281):1301-1315. [9] CHISĂLĂU BA, CRÎNGUȘ LI, VREJU FA, et al. New insights into IL‑17/IL‑23 signaling in ankylosing spondylitis. Exp Ther Med. 2020;20(4): 3493-3497. [10] GEORGESCU SR, TAMPA M, CARUNTU C, et al. Advances in understanding the immunological pathways in psoriasis. Int J Mol Sci. 2019;20(3): 739. [11] INGRASCIOTTA Y, SPINI A, L’ABBATE L, et al. Comparing clinical trial population representativeness to real-world users of 17 biologics approved for immune-mediated inflammatory diseases: an external validity analysis of 66,639 biologic users from the Italian VALORE project. Pharmacol Res. 2024; 200:107074. [12] BODUR H, ATAMAN Ş, BUĞDAYCI DS, et al. Description of the registry of patients with ankylosing spondylitis in Turkey: TRASD-IP. Rheumatol Int. 2012;32(1):169-176. [13] STOLWIJK C, VAN TUBERGEN A, CASTILLO-ORTIZ JD, et al. Prevalence of extra-articular manifestations in patients with ankylosing spondylitis: a systematic review and meta-analysis. Ann Rheum Dis. 2015;74(1):65-73. [14] GOTTLIEB AB, DEODHAR A, MCINNES IB, et al. Long-term safety of secukinumab over five years in patients with moderate-to-severe plaque psoriasis, psoriatic arthritis and ankylosing spondylitis: update on integrated pooled clinical trial and post-marketing surveillance data. Acta Derm Venereol. 2022; 102:adv00698. [15] MAN S, JI X, HU L, et al. Characteristics associated with the occurrence and development of acute anterior uveitis, inflammatory bowel disease, and psoriasis in patients with ankylosing spondylitis: data from the Chinese ankylosing spondylitis prospective imaging cohort. Rheumatol Ther. 2021;8(1):555-571. [16] CECCARELLI M, VENANZI RULLO E, BERRETTA M, et al. New generation biologics for the treatment of psoriasis and psoriatic arthritis. State of the art and considerations about the risk of infection. Dermatol Ther. 2021;34(1):e14660. [17] XU W D, LI R, HUANG A F. Role of TL1A in inflammatory autoimmune diseases: a comprehensive review. Front Immunol. 2022; 13:891328. [18] JUNG SM, KIM WU. Targeted immunotherapy for autoimmune disease. Immune Netw. 2022; 22(1):e9. [19] ATHANASSIOU L, KOSTOGLOU-ATHANASSIOU I, KOUTSILIERIS M, et al. Vitamin D and autoimmune rheumatic diseases. Biomolecules. 2023;13(4):709. [20] KRUEGER JG, WHARTON JR KA, SCHLITT T, et al. IL-17A inhibition by secukinumab induces early clinical, histopathologic, and molecular resolution of psoriasis. J Allergy Clin Immunol. 2019;144(3):750-763. [21] BRIDGEWOOD C, NEWTON D, BRAGAZZI N, et al. Unexpected connections of the IL-23/IL-17 and IL-4/IL-13 cytokine axes in inflammatory arthritis and enthesitis. Semin Immunol. 2021;58:101520. [22] TSUKAZAKI H, KAITO T. The role of the IL-23/IL-17 pathway in the pathogenesis of spondyloarthritis. Int J Mol Sci. 2020; 21(17):6401. [23] CHANDRASEKHARAN UM, KAUR R, HARVEY JE, et al. TNFR2 depletion reduces psoriatic inflammation in mice by downregulating specific dendritic cell populations in lymph nodes and inhibiting IL-23/IL-17 pathways. J Invest Dermatol. 2022;142(8):2159-2172.e9. [24] RAMIRO S, NIKIPHOROU E, SEPRIANO A, et al. ASAS-EULAR recommendations for the management of axial spondyloarthritis: 2022 update. Ann Rheum Dis. 2023;82(1):19-34. [25] TORRES T, PUIG L, VENDER R, et al. Drug survival of IL-12/23, IL-17 and IL-23 inhibitors for psoriasis treatment: a retrospective multi-country, multicentric cohort study. Am J Clin Dermatol. 2021;22(4):567-579. [26] BERGBOER JGM, OOSTVEEN AM, DE JAGER MEA, et al. Paediatric‐onset psoriasis is associated with ERAP1 and IL23R loci, LCE3C_LCE3B deletion and HLA‐C* 06. Br J Dermatol. 2012;167(4):922-925. [27] ROBERTS AR, APPLETON LH, CORTES A, et al. ERAP1 association with ankylosing spondylitis is attributable to common genotypes rather than rare haplotype combinations. Proc Natl Acad Sci U S A. 2017;114(3):558-561. [28] MOHAN KN. DNMT1: catalytic and non-catalytic roles in different biological processes. Epigenomics. 2022;14(10):629-643. [29] HAY AD, KESSLER NJ, GEBERT D, et al. Epigenetic inheritance is unfaithful at intermediately methylated CpG sites. Nat Commun. 2023;14(1):5336. [30] HU Y, XU B, HE J, et al. Hypermethylation of Smad7 in CD4+ T cells is associated with the disease activity of rheumatoid arthritis. Front Immunol. 2023;14:1104881. [31] YAN L, GENG Q, CAO Z, et al. Insights into DNMT1 and programmed cell death in diseases. Biomed Pharmacother. 2023;168: 115753. [32] REN Y. Regulatory mechanism and biological function of UHRF1–DNMT1-mediated DNA methylation. Funct Integr Genomics. 2022;22(6):1113-1126. [33] GARSHASBI M, MAHMOUDI M, RAZMARA E, et al. Identification of RELN variant p.(Ser2486Gly) in an Iranian family with ankylosing spondylitis; the first association of RELN and AS. Eur J Hum Genet. 2020;28(6):754-762. [34] ALFARDAN AS, NADEEM A, AHMAD SF, et al. DNMT inhibitor, 5-aza-2′-deoxycytidine mitigates di (2-ethylhexyl) phthalate-induced aggravation of psoriasiform inflammation in mice via reduction in global DNA methylation in dermal and peripheral compartments. Int Immunopharmacol. 2024;137:112503. [35] JIANG MM, ZHAO F, LOU TT. Assessment of significant pathway signaling and prognostic value of GNG11 in ovarian serous cystadenocarcinoma. Int J Gen Med. 2021;14:2329-2341. [36] MENG G, DUAN H, JIA J, et al. Alfalfa xenomiR-162 targets G protein subunit gamma 11 to regulate milk protein synthesis in bovine mammary epithelial cells. Anim Biosci. 2024;37(3):509-521. [37] ZHANG Y, ZHANG C, CHEN W, et al. The landscape of allelic expression and DNA methylation at the bovine SGCE/PEG10 locus. Anim Genet. 2024;55(3):452-456. [38] FANG F, PAN J, XU L, et al. Identification of potential transcriptomic markers in developing ankylosing spondylitis: a meta‐analysis of gene expression profiles. Biomed Res Int. 2015; 2015(1):826316. [39] WROŃSKI A, JAROCKA-KARPOWICZ I, STASIEWICZ A, et al. Phytocannabinoids in the pharmacotherapy of psoriasis. Molecules. 2023;28(3):1192. [40] CÁNOVAS PM. Survivin mediates mitotic onset in hela cells through activation of the Cdk1-Cdc25B axis. Res Sq. 2024;28: rs3949429. [41] FERENCOVA I, VASKOVICOVA M, DRUTOVIC D, et al. CDC25B is required for the metaphase I-metaphase II transition in mouse oocytes. J Cell Sci. 2022;135(6):jcs252924. [42] LI HX, YANG WY, LI LP, et al. Molecular dynamics study of CDC25BR492L mutant causing the activity decrease of CDC25B. J Mol Graph Model. 2021;109:108030. [43] GOETZKE CC, EBSTEIN F, KALLINICH T. Role of proteasomes in inflammation. J Clin Med. 2021;10(8):1783. [44] LI L P, LI H X, ZHOU H, et al. Exploring the mechanism of C473D mutation on CDC25B causing weak binding affinity with CDK2/CyclinA by molecular dynamics study. J Biomol Struct Dyn. 2023;41(22):12552-12564. [45] JARLBORG M, COURVOISIER DS, LAMACCHIA C, et al. Serum calprotectin: a promising biomarker in rheumatoid arthritis and axial spondyloarthritis. Arthritis Res Ther. 2020; 22(1):105. [46] PETIT RG, CANO A, ORTIZ A, et al. Psoriasis: from pathogenesis to pharmacological and nano-technological-based therapeutics. Int J Mol Sci. 2021;22(9):4983. [47] XIA P, JI X, YAN L, et al. Roles of S100A8, S100A9 and S100A12 in infection, inflammation and immunity. Immunology. 2024;171(3):365-376. [48] CERÓN JJ, ORTÍN-BUSTILLO A, LÓPEZ-MARTÍNEZ MJ, et al. S-100 proteins: basics and applications as biomarkers in animals with special focus on calgranulins (S100A8, A9, and A12). Biology. 2023;12(6):881. [49] SPRENKELER EGG, ZANDSTRA J, VAN KLEEF ND, et al. S100A8/A9 is a marker for the release of neutrophil extracellular traps and induces neutrophil activation. Cells. 2022;11(2):236. [50] VON WULFFEN M, LUEHRMANN V, ROBECK S, et al. S100A8/A9-alarmin promotes local myeloid-derived suppressor cell activation restricting severe autoimmune arthritis. Cell Rep. 2023;42(8):113006. [51] BORSKY P, FIALA Z, ANDRYS C, et al. Alarmins HMGB1, IL‐33, S100A7, and S100A12 in psoriasis vulgaris. Mediators Inflamm. 2020; 2020(1):8465083. [52] SINGH P, ALI SA. Multifunctional role of S100 protein family in the immune system: an update. Cells. 2022;11(15):2274. [53] ABDI W, ROMASCO A, ALKURDI D, et al. An overview of S100 proteins and their functions in skin homeostasis, interface dermatitis conditions and other skin pathologies. Exp Dermatol. 2024;33(8):e15158. [54] SREEJIT G, FLYNN MC, PATIL M, et al. S100 family proteins in inflammation and beyond. Adv Clin Chem. 2020;98:173-231. [55] LEŚNIAK W, FILIPEK A. S100 Proteins—intracellular and extracellular function in norm and pathology. Biomolecules. 2024;14(4):432. [56] SAGONAS I, ILIOPOULOS G, Baraliakos X, et al. Anti-TNF-α induced paradoxical psoriasis in patients with ankylosing spondylitis: a systematic review. Clin Exp Rheumatol. 2024; 42(1):178-184. [57] PAN S, WU S, WEI Y, et al. Exploring the causal relationship between inflammatory cytokines and inflammatory arthritis: a Mendelian randomization study. Cytokine. 2024;173:156446. [58] DEODHAR A, BLAUVELT A, LEBWOHL M, et al. Long-term safety of Ixekizumab in adults with psoriasis, psoriatic arthritis, or axial spondyloarthritis: a post-hoc analysis of final safety data from 25 randomized clinical trials. Arthritis Res Ther. 2024;26(1):49. [59] CARNAZZO V, REDI S, BASILE V, et al. Calprotectin: two sides of the same coin. Rheumatology. 2024; 63(1):26-33. [60] KAZAKOV AS, RASTRYGINA VA, VOLOGZHANNIKOVA AA, et al. Recognition of granulocyte-macrophage colony-stimulating factor by specific S100 proteins. Cell Calcium. 2024;119:102869. |
| [1] | Gao Zengjie, , Pu Xiang, Li Lailai, Chai Yihui, Huang Hua, Qin Yu. Increased risk of osteoporotic pathological fractures associated with sterol esters: evidence from IEU-GWAS and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1302-1310. |
| [2] | Liu Fengzhi, Dong Yuna, Tian Wenyi, Wang Chunlei, Liang Xiaodong, Bao Lin. Gene-predicted associations between 731 immune cell phenotypes and rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1311-1319. |
| [3] | Zhang Cuicui, Chen Huanyu, Yu Qiao, Huang Yuxuan, Yao Gengzhen, Zou Xu. Relationship between plasma proteins and pulmonary arterial hypertension and potential therapeutic targets [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1331-1340. |
| [4] | Zhang Qian, Huang Dongfeng. Weighted gene co-expression network analysis combined with machine learning to screen and validate biomarkers for osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1096-1105. |
| [5] | Li Guangzheng, Li Wei, Zhang Bochun, Ding Haoqin, Zhou Zhongqi, Li Gang, Liang Xuezhen. A prediction model for sarcopenia in postmenopausal women: information analysis based on the China Health and Retirement Longitudinal Study database [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 849-857. |
| [6] | Zeng Hao, Sun Pengcheng, Chai Yuan, Huang Yourong, Zhang Chi, Zhang Xiaoyun. Association between thyroid function and osteoporosis: genome-wide data analysis of European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1019-1027. |
| [7] | Rong Xiangbin, , Zheng Haibo, Mo Xueshen, Hou Kun, Zeng Ping, . Plasma metabolites, immune cells, and hip osteoarthritis: causal inference based on GWAS data from European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1028-1035. |
| [8] | He Qiwang, , , Chen Bo, Liang Fuchao, Kang Zewei, Zhou Yuan, Ji Anxu, Tang Xialin, . Relationship between Alzheimer’s disease and sarcopenia and body mass index: analysis of GWAS datasets for European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1036-1046. |
| [9] | Ding Yu, Chen Jingwen, Chen Xiuyan, Shi Huimin, Yang Yudie, Zhou Meiqi, Cui Shuai, . Circulating inflammatory proteins and myocardial hypertrophy: large sample analysis of European populations from GWAS Catalog and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1047-1057. |
| [10] | Gu Fucheng, Yang Meixin, Wu Weixin, Cai Weijun, Qin Yangyi, Sun Mingyi, Sun Jian, Geng Qiudong, Li Nan. Effects of Guilu Erxian Glue on gut microbiota in rats with knee osteoarthritis: machine learning and 16S rDNA analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1058-1072. |
| [11] | Guan Yujie, Zhao Bin. Application and prospect of artificial intelligence in screening and diagnosis of scoliosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 721-730. |
| [12] | Wang Zhipeng, Zhang Xiaogang, Zhang Hongwei, Zhao Xiyun, Li Yuanzhen, Guo Chenglong, Qin Daping, Ren Zhen. A systematic review of application value of machine learning to prognostic prediction models for patients with lumbar disc herniation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 740-748. |
| [13] | Liu Chu, Qiu Boyuan, Tong Siwen, He Linyuwei, Chen Haobo, Ou Zhixue. A genetic perspective reveals the relationship between blood metabolites and osteonecrosis: an analysis of information from the FinnGen database in Finland [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 785-794. |
| [14] | Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. The construction and validation of a prediction model based on multiple machine learning algorithms and the immunomodulatory analysis of rheumatoid arthritis from the perspective of mitophagy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-15. |
| [15] | Zhang Yibo, Lu Jianqi, Mao Meiling, Pang Yan, Dong Li, Yang Shangbing, Xiao Xiang. Exploring the causal relationship between rheumatoid arthritis and coronary atherosclerosis: a Mendel randomized study involving serum metabolites and inflammatory factors [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-9. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||