Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (8): 1741-1750.doi: 10.12307/2025.314
Previous Articles Next Articles
Bioinformatics analysis of potential biomarkers for primary osteoporosis
Zhao Jiacheng1, 2, Ren Shiqi3, Zhu Qin3, Liu Jiajia1, Zhu Xiang1, Yang Yang1, 2
- 1Department of Trauma Center, 3Department of Hand Surgery, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China; 2Medical School of Nantong University, Nantong 226001, Jiangsu Province, China
-
Received:2024-03-09Accepted:2024-04-09Online:2025-03-18Published:2024-07-06 -
Contact:Yang Yang, Master, Attending physician, Department of Trauma Center, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China; Medical School of Nantong University, Nantong 226001, Jiangsu Province, China -
About author:Zhao Jiacheng, Master candidate, Physician, Department of Trauma Center, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China; Medical School of Nantong University, Nantong 226001, Jiangsu Province, China -
Supported by:Jiangsu Research Hospital Project, No. YJXYY202204-YSB20 (to YY); Nantong Municipal Health Commission Science and Technology Project, No. MS2023016 (to YY); Jiangsu Postgraduate Research and Innovation Project, No. KYCX23-3433 (to ZJC)
CLC Number:
Cite this article
Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
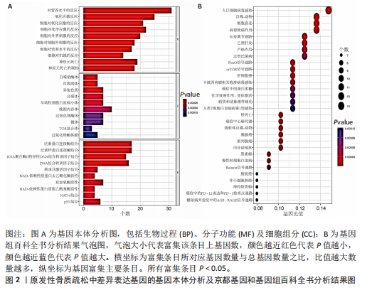
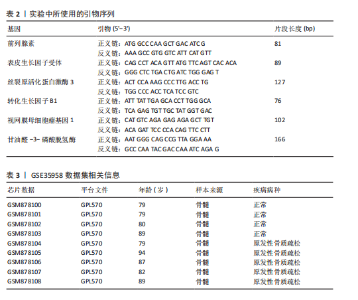
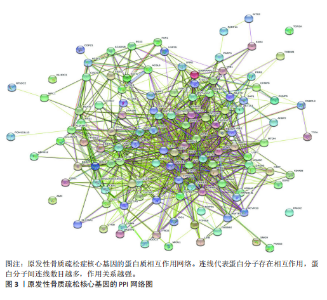
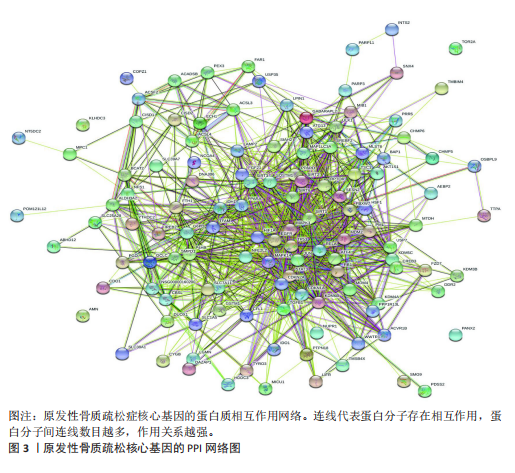
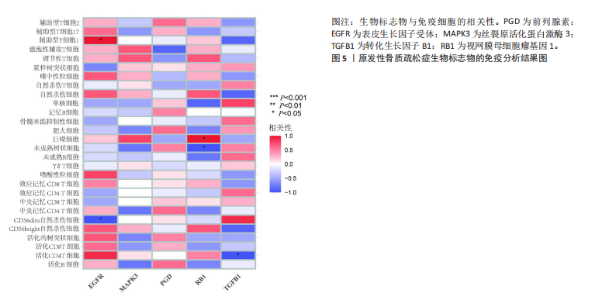
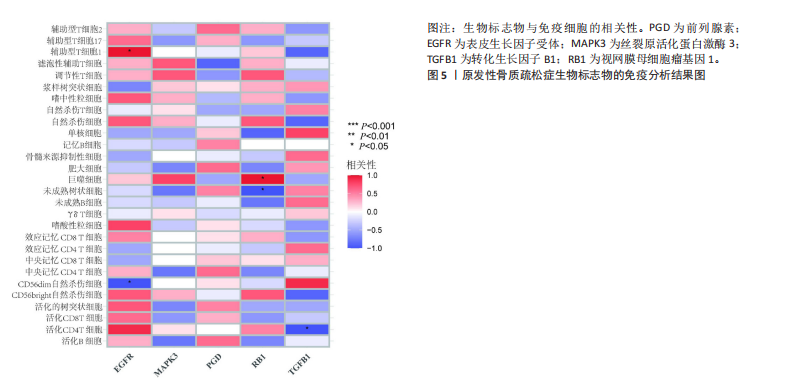
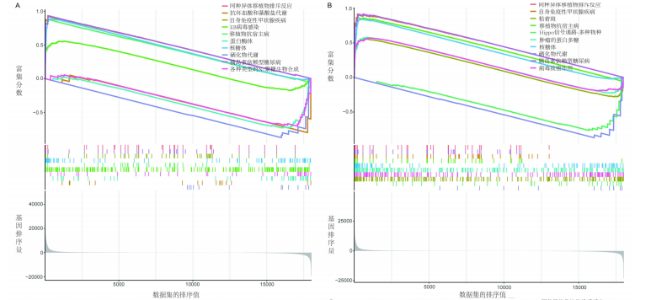
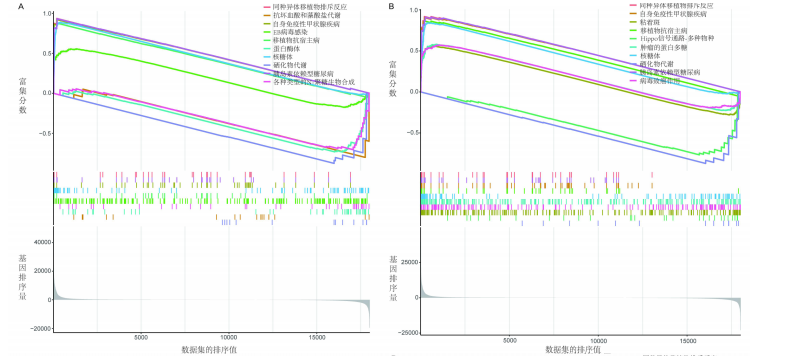
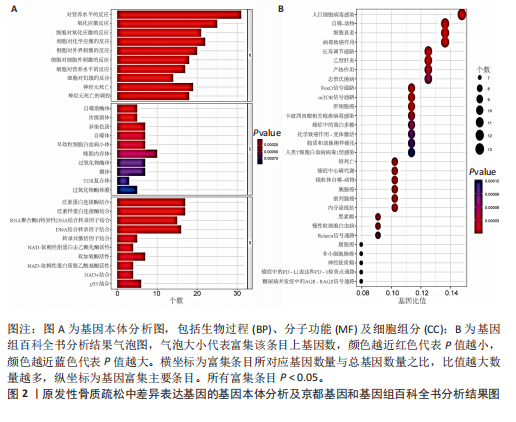
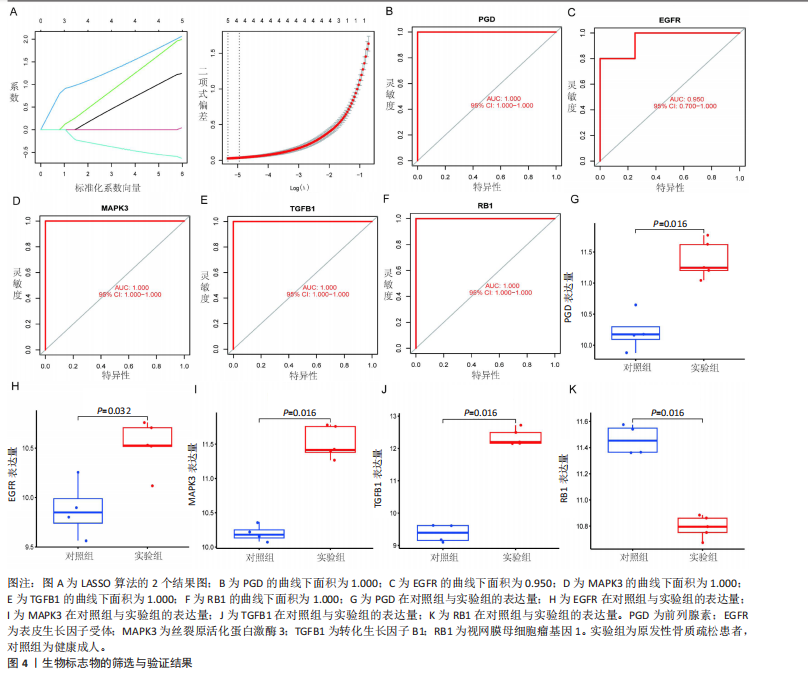
对细胞外刺激的反应、细胞对氧化应激的反应以及自噬的调节等生物过程中发挥作用,在细胞组分中,与自噬体、核被膜以及过氧物酶体等;在分子功能中,主要在DNA结合转录因子结合、泛素蛋白连接酶结合以及泛素样蛋白连接酶结合等发挥作用。京都基因和基因组百科全书的生物学机制分析显示,交集基因主要聚集在人巨细胞病毒感染、自噬以及细胞衰老等信号通路中。 2.3 蛋白质相互作用网络分析结果 使用STRING数据库构建了原发性骨质疏松中与核心差异基因相互作用的潜在蛋白质作用网络,见图3。结果中显示位于网络中心的核心基因影响其他基因在生物系统中的关系,共包括谷氨酸-半胱氨酸连接酶催化亚基、丝裂原活化蛋白激酶14、赖氨酸脱甲基酶6B及小鼠双微体2等27个核心基因,最后选取该网络中的27个核心基因进行生物标志物筛选。 2.4 生物标志物的筛选与验证结果 该研究通过最小绝对收缩选择算子逻辑回归算法从27个核心基因中鉴定出前列腺素、表皮生长因子受体、丝裂原活化蛋白激酶3、转化生长因子B1和视网膜母细胞瘤基因1等5个原发性骨质疏松相关的生物标志物,见图4A。 受试者工作特征分析曲线显示出它们具有较高的精确性,其中前列腺素的曲线下面积为1.000,表皮生长因子受体的曲线下面积为0.950,丝裂原活化蛋白激酶3的曲线下面积为1.000,转化生长因子B1的曲线下面积为1.000,以及视网膜母细胞瘤基因1的曲线下面积为1.000,见图4B-F。这些生物标志物在对照组即正常组织中与实验组即原发性骨质疏松组织中的差异表达分析显示,实验组中前列腺素、表皮生长因子受体、丝裂原活化蛋白激酶3以及转化生长因子B1的表达水平均明显高于对照组样本(P < 0.05),见图4G-J;而视网膜母细胞瘤基因1在实验组表达量明显低于对照组样本(P < 0.05),见图4K。 2.5 生物标志物的免疫分析结果 该研究中免疫分析结果表明,细胞免疫在原发性骨质疏松中起重要作用。通过CIBERSORT算法分析生物标志物在不同免疫细胞浸润情况,表皮生长因子受体与辅助型T细胞1密切关联,视网膜母细胞瘤基因1在巨噬细胞中显著聚集见图5。 2.6 生物标志物的基因富集分析结果 将原发性骨质疏松的生物标志物进行基因富集分析。前列腺素的分析结果表明,其高表达可能影响蛋白酶体和EB病毒感染等方面,低表达可能与抗坏血酸和醛达酸代谢等方面有关,见图6A。表皮生长因子受体的分析结果表明,其高表达可能影响同种异体移植排斥及自身免疫性甲状腺疾病等,低表达可能影响移植物抗宿主疾病等,见图6B。丝裂原活化蛋白激酶3的分析结果表明其与JAK?STAT信号通路、同种异体移植排斥及爱帕琳肽信号通路相关,见图6C。转化生长因子B1的分析结果表明,其与同种异体移植排斥、抗坏血酸和醛达酸代谢、自身免疫性甲状腺疾病等通路相"
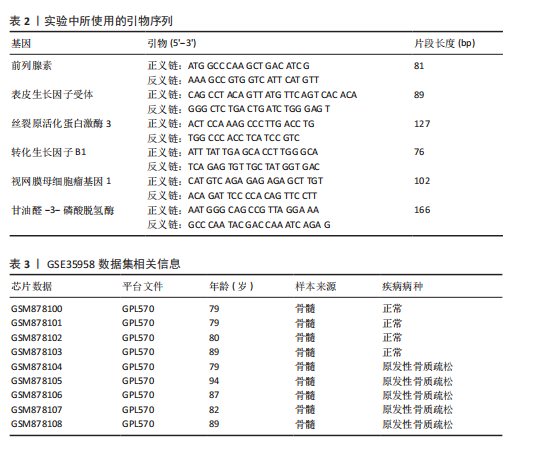
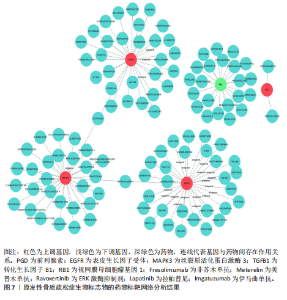
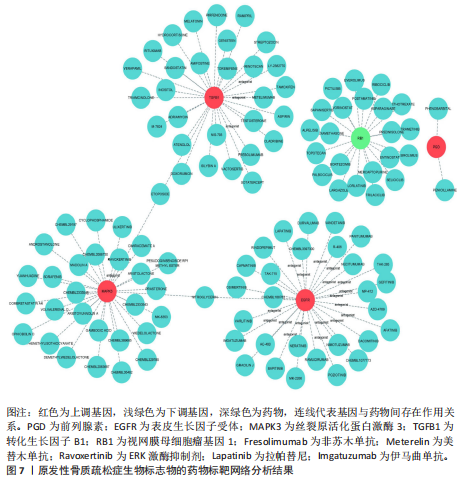
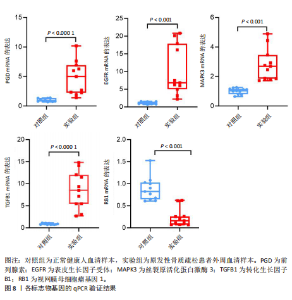
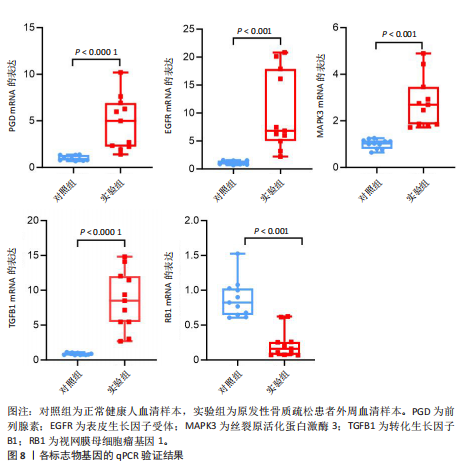
关,见图6D。视网膜母细胞瘤基因1的分析结果表明其与Ras相关蛋白1信号通路、同种异体移植排斥以及自身免疫性甲状腺疾病等显著相关,见图6E。 2.7 药物标靶网络分析结果 将获得的原发性骨质疏松生物标志物建立药物标靶网络,可以看到生物标志物与原发性骨质疏松相关药物基因的关系,见图7。其中转化生长因子B1与非苏木单抗、美替木单抗有协同作用;丝裂原活化蛋白激酶3与ERK激酶抑制剂Ravoxertinib存在协同作用;表皮生长因子受体与拉帕替尼、泛表皮生长因子受体小分子抑制剂以及伊马曲单抗等药物具有协同作用。 2.8 各标志物基因的qPCR验证结果 在以上生物信息学分析的基础上,收集原发性骨质疏松患者及正常健康人外周血清,提取RNA进行qPCR分析,结果表明前列腺素、表皮生长因子受体、丝裂原活化蛋白激酶3和转化生长因子B1在原发性骨质疏松患者的外周血清中转录水平上升(P < 0.001),见图8;而视网膜母细胞瘤基因1在原发性骨质疏松患者的外周血清中转录水平下降(P < 0.001),见图8。qPCR结果与文章的模型表现出良好的吻合度。"
| [1] LEBOFF MS, GREENSPAN SL, INSOGNA KL, et al. The clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2022;33(10):2049-2102. [2] BROWN JP. Long-term treatment of postmenopausal osteoporosis. Endocrinol Metab (Seoul). 2021;36(3):544-552. [3] 武瑞骐,周毅,夏天,等.类风湿性关节炎与骨质疏松症和骨密度关系的孟德尔随机化分析[J].中国组织工程研究,2024,28(23): 3715-3721. [4] YONG EL, LOGAN S. Menopausal osteoporosis: screening, prevention and treatment. Singapore Med J. 2021;62(4):159-166. [5] LI S, CHEN B, CHEN H, et al. Analysis of potential genetic biomarkers and molecular mechanism of smoking-related postmenopausal osteoporosis using weighted gene co-expression network analysis and machine learning. PLoS One. 2021;16(9): e0257343. [6] AIBAR-ALMAZAN A, VOLTES-MARTINEZ A, CASTELLOTE-CABALLERO Y, et al. Current status of the diagnosis and management of osteoporosis. Int J Mol Sci. 2022;23(16):9465. [7] HU Y, HAN J, DING S, et al. Identification of ferroptosis-associated biomarkers for the potential diagnosis and treatment of postmenopausal osteoporosis. Front Endocrinol (Lausanne). 2022;13:986384. [8] DEMPSTER DW, SHANE E, HORBERT W, et al. A simple method for correlative light and scanning electron microscopy of human iliac crest bone biopsies: qualitative observations in normal and osteoporotic subjects. J Bone Miner Res. 1986;1(1):15-21. [9] RIGGS BL, KHOSLA S, ATKINSON EJ, et al. Evidence that type I osteoporosis results from enhanced responsiveness of bone to estrogen deficiency. Osteoporos Int. 2003;14(9):728-733. [10] SRIVASTAVA M, DEAL C. Osteoporosis in elderly: prevention and treatment. Clin Geriatr Med. 2002;18(3):529-555. [11] LI Y, SI Y, MA Y, et al. Application and prospect of metabolomics in the early diagnosis of osteoporosis: a narrative review. Bioanalysis. 2023;15(22):1369-79. [12] PRESTWOOD KM, PILBEAM CC, RAISZ LG. Treatment of osteoporosis. Annu Rev Med. 1995;46:249-256. [13] CHEUNG AM, GIANGREGORIO L. Mechanical stimuli and bone health: what is the evidence?. Curr Opin Rheumatol. 2012;24(5):561-566. [14] WANG CG, HU YH, SU SL, et al. LncRNA DANCR and miR-320a suppressed osteogenic differentiation in osteoporosis by directly inhibiting the Wnt/beta-catenin signaling pathway. Exp Mol Med. 2020;52(8):1310-1325. [15] YOSHIDA G, KAWABATA T, TAKAMATSU H, et al. Degradation of the NOTCH intracellular domain by elevated autophagy in osteoblasts promotes osteoblast differentiation and alleviates osteoporosis. Autophagy. 2022;18(10):2323-2332. [16] HUANG H, HE YM, LIN MM, et al. P2X7Rs: new therapeutic targets for osteoporosis. Purinergic Signal. 2023;19(1):207-219. [17] AYERS C, KANSAGARA D, LAZUR B, et al. Effectiveness and safety of treatments to prevent fractures in people with low bone mass or primary osteoporosis: a living systematic review and network meta-analysis for the american college of physicians. Ann Intern Med. 2023;176(2):182-195. [18] KIM W K, MELITON V, BOURQUARD N, et al. Hedgehog signaling and osteogenic differentiation in multipotent bone marrow stromal cells are inhibited by oxidative stress. J Cell Biochem. 2010;111(5):1199-1209. [19] H DEHKORD IM, TASHAKOR A, O’CONNELL E, et al. Apoptosome-dependent myotube formation involves activation of caspase-3 in differentiating myoblasts. Cell Death Dis. 2020;11(5):308. [20] LI F, SUN X, MA J, et al. Naringin prevents ovariectomy-induced osteoporosis and promotes osteoclasts apoptosis through the mitochondria-mediated apoptosis pathway. Biochem Biophys Res Commun. 2014;452(3):629-635. [21] ZHANG C, LI H, LI J, et al. Oxidative stress: a common pathological state in a high-risk population for osteoporosis. Biomed Pharmacother. 2023;163:114834. [22] SANCHEZ-RODRIGUEZ MA, RUIZ-RAMOS M, CORREA-MUNOZ E, et al. Oxidative stress as a risk factor for osteoporosis in elderly Mexicans as characterized by antioxidant enzymes. BMC Musculoskelet Disord. 2007;8:124. [23] LIU Y, WANG C, WANG G, et al. Loureirin B suppresses RANKL-induced osteoclastogenesis and ovariectomized osteoporosis via attenuating NFATc1 and ROS activities. Theranostics. 2019;9(16):4648-4662. [24] LI H, HUANG C, ZHU J, et al. Lutein suppresses oxidative stress and inflammation by nrf2 activation in an osteoporosis rat model. Med Sci Monit. 2018;24:5071-5075. [25] RAUWEL B, DEGBOE Y, DIALLO K, et al. Inhibition of osteoclastogenesis by the RNA-binding protein QKI5: a novel approach to protect from bone resorption. J Bone Miner Res. 2020;35(4):753-765. [26] CHANDRA A, LAGNADO AB, FARR JN, et al. Targeted reduction of senescent cell burden alleviates focal radiotherapy-related bone loss. J Bone Miner Res. 2020;35(6):1119-1131. [27] FARR JN, XU M, WEIVODA MM, et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med. 2017;23(9):1072-10729. [28] GUAN JL, SIMON AK, PRESCOTT M, et al. Autophagy in stem cells. Autophagy. 2013;9(6): 830-849. [29] LI H, LI D, MA Z, et al. Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy. 2018;14(10):1726-1741. [30] YIN X, ZHOU C, LI J, et al. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res. 2019;7:28. [31] SHAPIRO IM, LAYFIELD R, LOTZ M, et al. Boning up on autophagy: the role of autophagy in skeletal biology. Autophagy. 2014;10(1):7-19. [32] ZHU C, SHEN S, ZHANG S, et al. Autophagy in bone remodeling: a regulator of oxidative stress. Front Endocrinol (Lausanne). 2022;13: 898634. [33] WU M, ZHANG P. EGFR-mediated autophagy in tumourigenesis and therapeutic resistance. Cancer Lett. 2020;469:207-216. [34] LIANG C, XU J, MENG Q, et al. TGFB1-induced autophagy affects the pattern of pancreatic cancer progression in distinct ways depending on SMAD4 status. Autophagy. 2020;16(3):486-500. [35] XIAO S, LIU N, YANG X, et al. Polygalacin D suppresses esophageal squamous cell carcinoma growth and metastasis through regulating miR-142-5p/Nrf2 axis. Free Radic Biol Med. 2021;164:58-75. [36] RUBY M, GIFFORD CC, PANDEY R, et al. Autophagy as a therapeutic target for chronic kidney disease and the roles of TGF-beta1 in autophagy and kidney fibrosis. Cells. 2023. doi:10.3390/cells12030412. [37] TIAN L, XIAO H, LI M, et al. A novel Sprouty4-ERK1/2-Wnt/beta-catenin regulatory loop in marrow stromal progenitor cells controls osteogenic and adipogenic differentiation. Metabolism. 2020;105:154189. [38] LIU F, SONG C, CAI W, et al. Shared mechanisms and crosstalk of COVID-19 and osteoporosis via vitamin D. Sci Rep. 2022;12(1):18147. [39] LINDER M, HECKING M, GLITZNER E, et al. EGFR controls bone development by negatively regulating mTOR-signaling during osteoblast differentiation. Cell Death Differ. 2018;25(6):1094-106. [40] ROBLING AG, BONEWALD LF. The osteocyte: new insights. Annu Rev Physiol. 2020;82:485-506. [41] YUE J, LOPEZ J M. Understanding MAPK signaling pathways in apoptosis. Int J Mol Sci. 2020;21(7):2346. [42] TANG L, WU M, LU S, et al. Fgf9 Negatively regulates bone mass by inhibiting osteogenesis and promoting osteoclastogenesis Via MAPK and PI3K/AKT signaling. J Bone Miner Res. 2021;36(4):779-791. [43] 覃浩,亢腾,刘钢.长链非编码RNA通过p38MAPK信号通路直接或间接影响骨质疏松症[J].中国组织工程研究,2025,29(1):175-184. [44] LIU Q, XU YC, SHUAI ZW. Mir-142-3P regulates MAPK protein family by inhibiting 14-3-3η to enhance bone marrow mesenchymal stem cells osteogenesis. Sci Rep. 2023;13(1):22862. [45] TAKAHARA T, TSUYUKI T, SATOU A, et al. TGFB1 mRNA expression is associated with poor prognosis and specific features of inflammation in ccRCC. Virchows Arch. 2022;480(3):635-643. [46] WANG YW, LIN WY, WU FJ, et al. Unveiling the transcriptomic landscape and the potential antagonist feedback mechanisms of TGF-beta superfamily signaling module in bone and osteoporosis. Cell Commun Signal. 2022; 20(1):190. [47] SASAKI N, ITAKURA Y, MOHSIN S, et al. Cell surface and functional features of cortical bone stem cells. Int J Mol Sci. 2021;22(21):11849. [48] TAKEUCHI Y, NAKAYAMA K, MATSUMOTO T. Differentiation and cell surface expression of transforming growth factor-beta receptors are regulated by interaction with matrix collagen in murine osteoblastic cells. J Biol Chem. 1996; 271(7):3938-3944. [49] KNUDSEN ES, PRUITT SC, HERSHBERGER PA, et al. Cell cycle and beyond: exploiting new RB1 controlled mechanisms for cancer therapy. Trends Cancer. 2019;5(5):308-324. [50] THOMAS DM, CARTY SA, PISCOPO DM, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8(2):303-216. [51] CALO E, QUINTERO-ESTADES JA, DANIELIAN PS, et al. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010; 466(7310):1110-1114. [52] HANAU S, HELLIWELL JR. 6-Phosphogluconate dehydrogenase and its crystal structures. Acta Crystallogr F Struct Biol Commun. 2022; 78(Pt 3):96-112. |
| [1] | Lai Jiaming, , Song Yuling, Chen Zixi, Wei Jinghuan, Cai Hao, , Li Guoquan, . Screening of diagnostic markers for endothelial cell Senescence in mice with radiation-induced heart disease and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1450-1463. |
| [2] | Lyu Guoqing, Aizimaitijiang·Rouzi, Xiong Daohai. Irisin inhibits ferroptosis in human articular chondrocytes: roles and mechanisms [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1359-1367. |
| [3] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [4] | Wang Zhengye, Liu Wanlin, Zhao Zhenqun. Advance in the mechanisms underlying miRNAs in steroid-induced osteonecrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1207-1214. |
| [5] | Yu Daiyao, Shi Ping, Yang Lan, Li Zhishu, Lu Yongping. Advances and application of neutrophil extracellular traps and activated platelets in lung cancer research [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(1): 229-237. |
| [6] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [7] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [8] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [9] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [10] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [11] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [12] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [13] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| [14] | Jiang Qiyu, Zeng Huiyan. A novel analysis and prediction method for potential mechanisms of traditional Chinese medicine based on artificial intelligence and omics data-driven approach [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7552-7561. |
| [15] | Chen Ying, Guo Xiaojing, Mo Xueni, Ma Wei, Wu Shangzhi, Li Xiangling, Xie Tingting. Analysis of diagnostic biomarkers for ischemic stroke and experimental validation of targeted cuproptosis related genes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7562-7570. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||