Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (35): 7552-7561.doi: 10.12307/2025.968
Previous Articles Next Articles
A novel analysis and prediction method for potential mechanisms of traditional Chinese medicine based on artificial intelligence and omics data-driven approach
Jiang Qiyu1, Zeng Huiyan2
- 1Guangzhou University of Chinese Medicine, Guangzhou 510006, Guangdong Province, China; 2Guangdong Provincial Hospital of Traditional Chinese Medicine, Guangzhou 510120, Guangdong Province, China
-
Received:2024-11-15Accepted:2024-12-23Online:2025-12-18Published:2025-05-06 -
Contact:Zeng Huiyan, MD, Chief physician, Guangdong Provincial Hospital of Traditional Chinese Medicine, Guangzhou 510120, Guangdong Province, China -
About author:Jiang Qiyu, Master, Lecturer, Guangzhou University of Chinese Medicine, Guangzhou 510006, Guangdong Province, China -
Supported by:National Natural Science Foundation of China, No. 82374233 (to ZHY); Natural Science Foundation of Guangdong Province, No. 2414050003181 (to ZHY)
CLC Number:
Cite this article
Jiang Qiyu, Zeng Huiyan. A novel analysis and prediction method for potential mechanisms of traditional Chinese medicine based on artificial intelligence and omics data-driven approach[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7552-7561.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
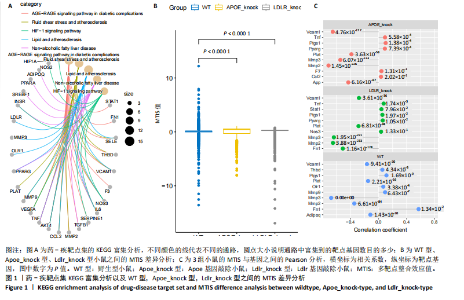
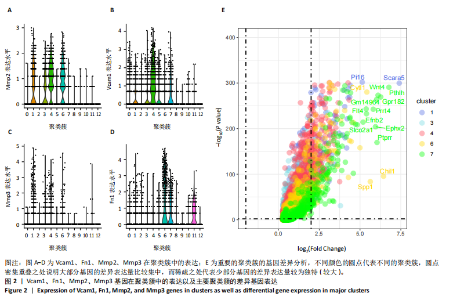
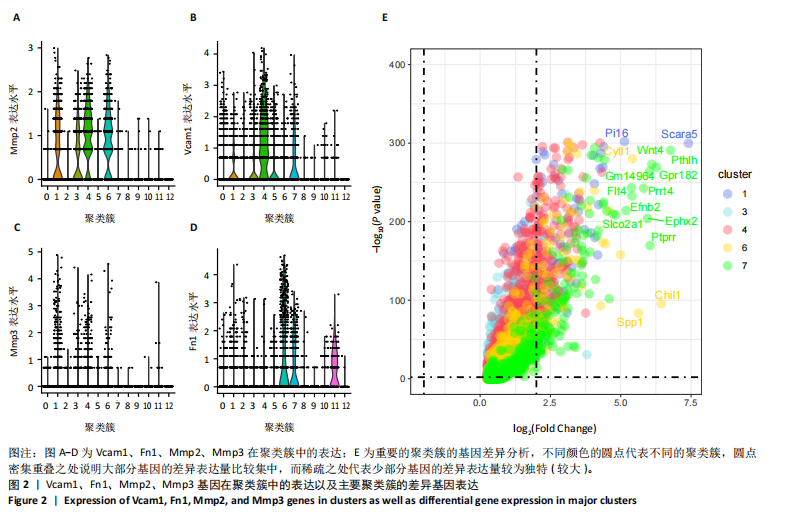
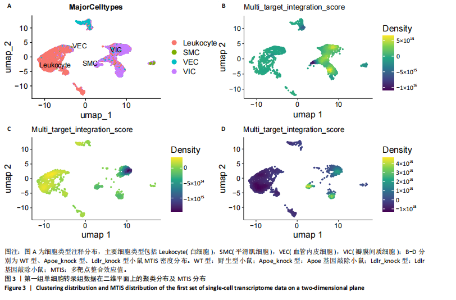
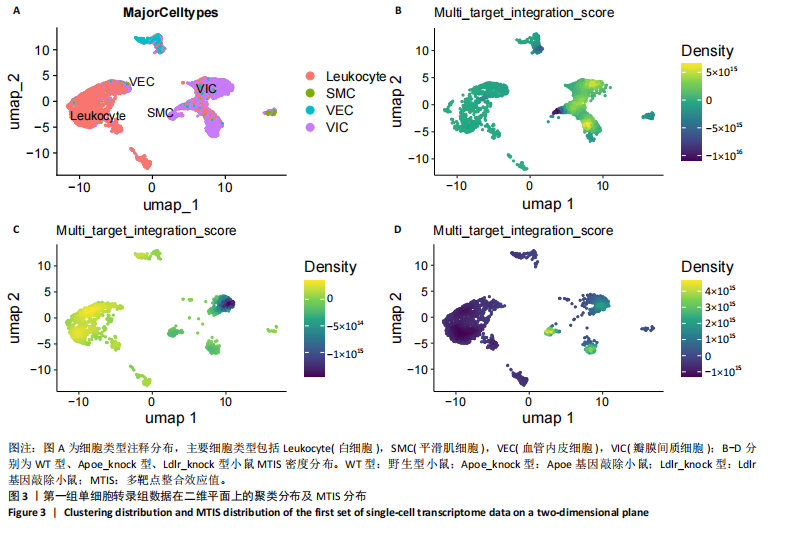
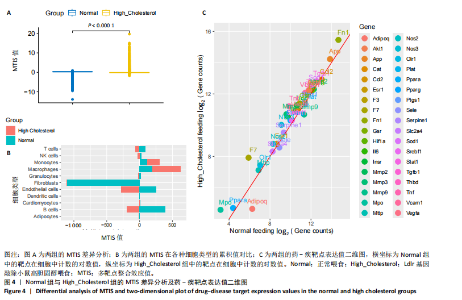
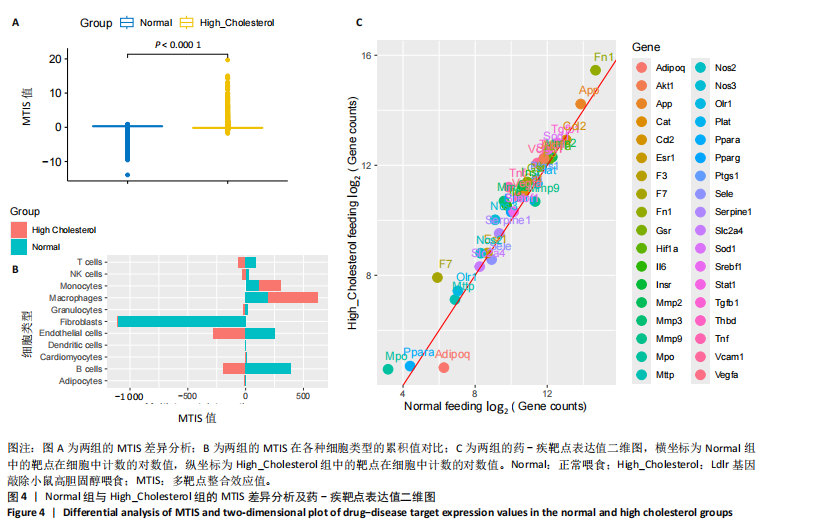
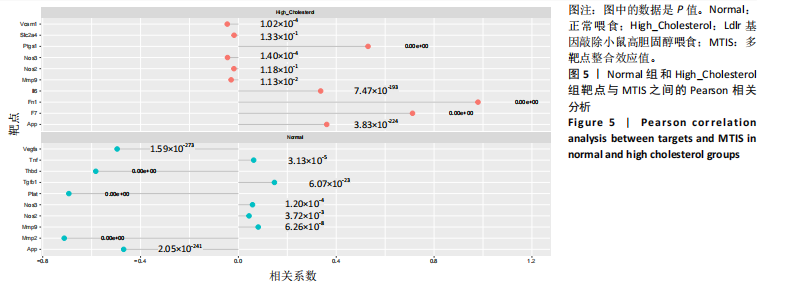
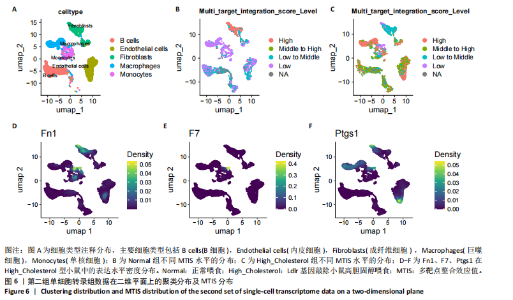
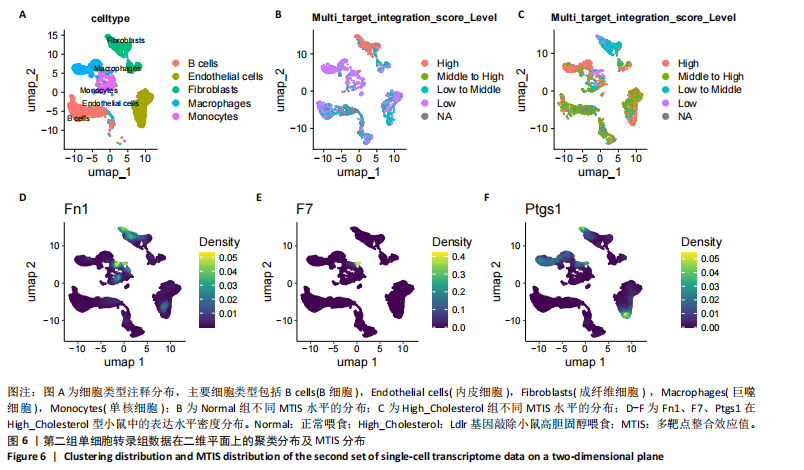
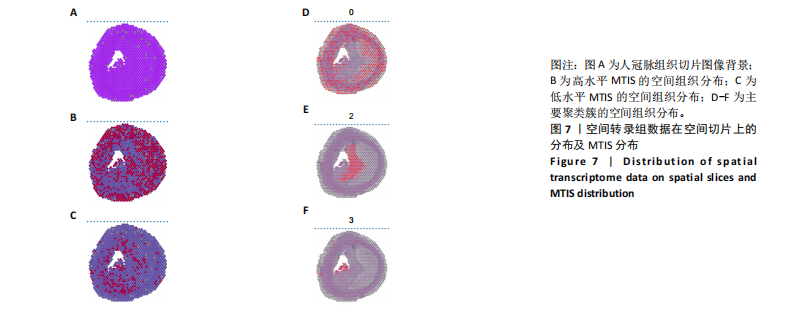
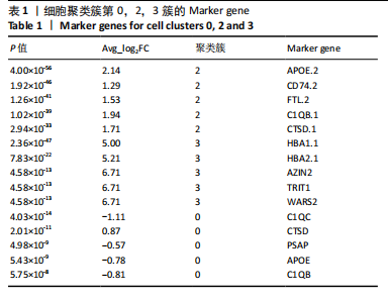
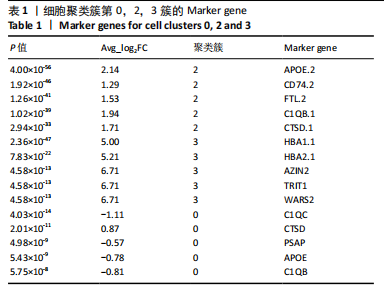
2.1 第一组单细胞转录组数据分析结果 2.1.1 药-疾靶点集KEGG富集分析 研究获取的药-疾靶点集包括LDLR,APOB,CCL2,MMP3,VEGFA,IL6,VCAM1,PTGS1,VEGFA等40余个基因。图1A所示,对药-疾靶点集进行KEGG富集处理,获取其中P < 0.01的前5位通路,具体包括AGE-RAGE通路、Fluid shear stress and atherosclerosis通路、HIF-1通路、Lipids and atherosclerosis通路以及Non?alcoholic fatty liver disease通路。除了HIF-1通路外,AGE-RAGE通路与糖尿病糖脂代谢有关,其余3条通路均与高血脂、动脉粥样硬化以及脂肪肝有关,这说明收集的药-疾靶点集是大柴胡汤治疗高脂血症以及动脉粥样硬化的代表性靶点,这些靶点的整合效应能够反映大柴胡汤的治疗作用。 2.1.2 MTIS比较分析 基于正常组(WT型)与异常组(基因敲除)之间的MTIS差异推测大柴胡汤对高脂血症的潜在机制推理过程如下:由于实验和临床已经证明大柴胡汤对高脂血症有作用是事实,而MTIS是药物和疾病多个共同靶点的整合映射值,MTIS差异的本质是药物与疾病共性靶点在正常组和疾病组中的表达水平差异,因此MTIS的变化在一定程度上反映了药物对疾病的潜在机制。如果在3组小鼠MTIS的均值对比中,正常组的MTIS值比其余两组的MTIS值要低,那么可以推测,高MTIS值为异常值,大柴胡汤此时的作用应该在于降低MTIS值,因此大柴胡汤的作用可能以MTIS负性效应为主,也就是说,大柴胡汤的潜在机制在此时应该是激活MTIS负相关的靶基因,或者抑制MTIS正相关的靶基因,当然也不能排除同时存在一些相反的作用,但应该不是大柴胡汤此时的主要效应。反之,如果在3组MTIS的均值对比中,正常组的MTIS值比其余两组的MTIS值要高,那么可以推测,低MTIS值为异常值,此时大柴胡汤的作用可能以MTIS正性效应为主,大柴胡汤的潜在机制在此时应该是激活MTIS正相关的靶基因,或者抑制MTIS负相关的靶基因。 将第一组3个样本单细胞转录组数据与药-疾靶点集的基因交集表达数据分别输入deep count autoencoder network后为每个细胞生成MTIS并进行统计学检验,结果如图1B所示,WT型与Apoe_knock型之间的MTIS值存在数据形态以及统计学上的差异(P < 0.000 1),WT型与Ldlr_knock型之间的MTIS值亦存在数据形态以及统计学上的差异(P < 0.000 1)。 在图1B中,WT型的MTIS正负数据基本平衡,这说明WT型的不同细胞中对药-疾靶点集的表达量各有差异,而且比较平均,这可能提示大柴胡汤对WT型的正常小鼠没有明显的效应。而相对于WT型而言,Ldlr_knock型和Apoe_knock型的数据形态与WT型明显不同,Ldlr_knock型和Apoe_knock型的MTIS均值均比WT组小,这表明大柴胡汤对Ldlr_knock型和Apoe_knock型小鼠的作用以MTIS正性效应为主,大柴胡汤对Ldlr_knock型和Apoe_knock型的潜在机制在此时应该是激活MTIS正相关的靶基因,或者抑制MTIS负相关的靶基因。 2.1.3 潜在效应基因 图1C展示了与3种类型小鼠MTIS之间的主要相关基因,相对于WT型,Apoe_knock型中MTIS负相关系数增加的基因是Vcam1,而Ldlr_knock型中MTIS负相关系数显著增加的是Fn1。Ldlr_knock型和Apoe_knock型两组中MTIS负相关系数均有明显增加的是Mmp2。与WT型对比,其余两组似乎没有MTIS正相关系数明显增加的基因。因此,大柴胡汤对于Ldlr_knock型和Apoe_knock型的潜在效应基因可能是Vcam1,Fn1,Mmp2,并且可能以抑制效应(对上述基因负调控作用)为主。 2.1.4 聚类簇差异基因表达 使用FindClusters算法聚类后通过VlnPloth函数观察基因Vcam1、Fn1、Mmp2、Mmp3在各聚类簇上的表达情况。根据图2A-D上4个基因在不同聚类簇上表达进一步表明,Vcam1和Mmp2在簇4中均有比较明显的表达,也就是说Vcam1和Mmp2可能存在直接或间接的协同调控作用。而3个基因至少其中2个在第1,3,4,6,7簇都有交集,图2E展示了图2A-D交集簇的差异表达基因,例如Flt4基因为血管内皮生长因子调控基因,参与动脉粥样硬化。 2.1.5 潜在效应细胞 经过UMAP算法降维处理后,第一组单细胞转录组数据在二维平面上的聚类分布如图3所示,图3A为主要细胞类型的分布,包括白细胞、平滑肌细胞、血管内皮细胞以及主动脉瓣间质细胞。图3B-D为3种类型小鼠的MTIS密度分布,可知3种类型的MTIS分布是明显不同的。与WT型小鼠的MTIS密度分布对比,Apoe_knock型小鼠的暗色区域与图3A中主动脉瓣间质细胞的区域重合,而在WT型小鼠中该区域是光亮的,表明Apoe_knock型小鼠在主动脉瓣间质细胞区域暗淡了。由于大柴胡汤对Apoe_knock型小鼠和WT型小鼠的MTIS在主动脉瓣间质细胞存在差异,而MTIS差异的本质是药物与疾病共性靶点在正常组和疾病组中的表达水平差异,因此大柴胡汤对Apoe_knock型小鼠的潜在效应细胞可能是主动脉瓣间质细胞。同理,Ldlr_knock型小鼠的暗色区域与白细胞、平滑肌细胞、血管内皮细胞的区域重合,说明大柴胡汤对Ldlr_knock型小鼠的潜在效应可能主要作用于这3种细胞。 2.2 第二组单细胞转录组数据分析结果 2.2.1 MTIS比较分析 见图4。High_Cholesterol组的靶基因原始表达明显高于Normal组,这说明MTIS对数据形态的映射是正确的。将第二组两个样本单细胞转录组数据与药-疾靶点集的基因交集表达数据分别输入deep count autoencoder network后为每个细胞生成MTIS并进行统计学检验,结果如图4所示,Ldlr基因敲除小鼠高胆固醇喂食(High_Cholesterol)与正常喂食(Normal)的MTIS存在差异,其数据形态明显不同,High_Cholesterol组的MTIS均值高于Normal组(P < 0.000 1)。 2.2.2 潜在效应基因 根据上述基于正常组与异常组之间的MTIS差异推测大柴胡汤对高脂血症的潜在机制的推理方法,可知相对于Normal型小鼠而言,大柴胡汤对High_Cholesterol型小鼠的作用以MTIS负性效应为主,在此时潜在机制是抑制MTIS正相关的靶基因,或者激活MTIS负相关的靶基因。在图5中,两组靶点与MTIS之间的Pearson相关分析对比表明,相对于Normal型小鼠,在High_Cholesterol型小鼠中,Fn1、F7、Ptgs1、IL6,App与MTIS正相关系数显著增加,但High_Cholesterol型似乎没有负相关系数明显增加的基因。因此可以推测,大柴胡汤对于High_Cholesterol型的潜在效应基因可能是Fn1、F7、Ptgs1、IL6,App,并且可能以抑制效应(对上述基因负调控作用)为主。它们与细胞黏附,凝血以及炎症过程有关。 2.2.3 潜在效应细胞 两组的MTIS在各种细胞类型的累积值对比,可见High_Cholesterol型的MTIS正性累积值占主导地位的是巨噬细胞,而MTIS差异的本质是药物与疾病共性靶点在正常组和疾病组中的表达水平差异,这表明大柴胡汤对High_Cholesterol型小鼠的潜在效应可能主要集中于巨噬细胞,可能涉及免疫反应过程。 在经过UMAP算法降维处理后,第二组单细胞转录组数据在二维平面上的聚类分布如图6所示。相对于Normal型小鼠MTIS的分布(图6B),High_Cholesterol型小鼠MTIS高值区域(图6C)与图6A的巨噬细胞区域重合,印证了巨噬细胞可能是大柴胡汤对High_Cholesterol型小鼠的效应细胞。从图6D-F观察可得知,Fn1、F7、Ptgs1三个基因的共性密度亮区亦出现在巨噬细胞的部分区域中,再次佐证了上述推测。因此,大柴胡汤对Ldlr基因敲除小鼠高胆固醇喂食的潜在效应可能与血管内皮损伤以及免疫介导有关。 经过上述分析可知,大柴胡汤治疗高脂血症的关键效应基因包括Fn1、Vcam1、Mmp2、IL6、Ptgs1,App等。大柴胡汤的潜在效应基因表明其机制可能主要以抗血管内皮细胞黏附,血管内皮炎症以及免疫反应调控为主。 2.3 空间转录组数据分析 将空间转录组细胞测序数据与药-疾靶点集的基因交集表达数据输入deep count autoencoder network后为每个细胞生成MTIS。 2.3.1 MTIS分布 冠脉组织的空间转录组MTIS对比如图7所示,MTIS高值细胞似乎在血管以及硬化斑块区域都有分布,但在血管区域的密度稍高,而MTIS低值细胞似乎主要集中于血管内皮以及硬化斑块区域等病变区域。这说明大柴胡汤对人类冠脉粥样硬化的潜在作用可能以调控MTIS低值细胞的靶基因为主。由于病变血管的斑块主要由吞噬了低密度脂蛋白的泡沫细胞侵袭到血管内皮并进一步导致内膜纤维组织增生及变性坏死而形成,因此可以推测MTIS低值细胞中的靶基因可能与泡沫细胞聚集到血管内皮,血管内膜纤维增生,纤维斑块变性坏死等病理过程有关,大柴胡汤通过调控这些病理过程,或许可减轻动脉粥样硬化的进程。 2.3.2 聚类簇的空间分布 使用FindClusters算法确定细胞聚类簇后,进一步探索了主要的细胞聚类簇在冠脉切片上的空间分布情况,如图7所示。图7D-F展示了细胞聚类簇第0,2,3簇的细胞在冠脉切片上的空间分布情况,可见第2簇和第3簇主要集中在硬化斑块区域。这些聚类簇的前5位Marker基因(差异基因)如表1所示,第2簇和第3簇的Marker基因为APOE.2、CD74.2、FTL.2、C1QB.1、CTSD.1、HBA1.1、HBA2.1、AZIN2、TRIT1、WARS2、FAM20B。由于实验已证实大柴胡汤对动脉粥样硬化存在一定的作用,且上述表明MTIS低值细胞似乎主要集中于血管内皮以及硬化斑块区域等病变区域,而这些Marker基因也主要集中于硬化斑块区域,因此这些Marker基因与大柴胡汤部分靶点可能存在一定的联系,但具体存在何种相互作用值得进一步探索。"
| [1] 王志杰,樊薛津,王豫骞,等.机器学习方法在中医药研究中的应用进展[J].药物评价研究,2024,47(8):1906-1913. [2] 邓乐,丁长松,黄辛迪,等.基于多层前馈神经网络的中药药性量化研究[J].中草药,2020,51(16):4277-4283. [3] 姜皓,张冰,张晓朦,等.基于4种机器学习算法的妊娠期中药“禁忌慎”判别[J].中草药,2021,52(24):7596-7605. [4] CHEN C, LI SX, WANG SM, et al. A support vector machine based pharmacodynamic prediction model for searching active fraction and ingredients of herbal medicine: Naodesheng prescription as an example. J Pharm Biomed Anal. 2011;56(2):443-447. [5] 王宁.基于异质网络的中药靶点预测及应用研究[D].北京:北京交通大学, 2022. [6] ZHANG P, ZHANG D, ZHOU W, et al. Network pharmacology: towards the artificial intelligence-based precision traditional Chinese medicine. Brief Bioinform. 2023; 25(1):bbad518. [7] 杨鹏挥,金丽君,廖杰,等.基于单细胞组学的中药现代研究:技术及思路[J].中国中药杂志,2022,47(15):3977-3985. [8] 陈嘉鋆,郭秋岩,徐承超,等.中药现代化研究的崭新模式:单细胞药理学[J].药学学报,2021,56(12):3300-3312. [9] RU J, LI P, WANG J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [10] STELZER G, ROSEN N, PLASCHKES I, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [11] GEER LY, MARCHLER-BAUER A, GEER RC, et al. The NCBI BioSystems database.Nucleic Acids Res. 2010;38(Database issue):D492-496. [12] ZHOU Y, ZHANG Y, ZHAO D, et al. TTD:Therapeutic Target Database describing target druggability information. Nucleic Acids Res. 2024;52(D1):D1465-D1477. [13] 梁天坚,李黎明,李本杰,等.基于网络药理学分析大柴胡汤治疗高脂血症的作用机制[J].中成药,2021,43(6):1645-1652. [14] LEE SH, KIM N, KIM M, et al. Single-cell transcriptomics reveal cellular diversity of aortic valve and the immunomodulation by PPARγ during hyperlipidemia.Nat Commun. 2022;13(1):5461. [15] VAN KUIJK K, MCCRACKEN IR, TILLIE RJHA, et al. Human and murine fibroblast single-cell transcriptomics reveals fibroblast clusters are differentially affected by ageing and serum cholesterol. Cardiovasc Res. 2023;119(7):1509-1523. [16] KAWAI K, SAKAMOTO A, MOKRY M, et al. Clonal Proliferation Within Smooth Muscle Cells in Unstable Human Atherosclerotic Lesions. Arterioscler Thromb Vasc Biol. 2023;43(12):2333-2347. [17] ERASLAN G, SIMON LM, MIRCEA M, et al. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10(1):390. [18] BAHL E, CHATTERJEE S, MUKHERJEE U, et al. Using deep learning to quantify neuronal activation from single-cell and spatial transcriptomic data. Nat Commun. 2024;15(1):779. [19] 刘恺.大柴胡汤治疗高脂血症的临床研究[J].内蒙古中医药,2023,42(8):82-84. [20] 宋伟.大柴胡汤治疗高脂血症50例[J].实用中医药杂志,2022,38(5):735-736. [21] 杨荣来,王凤荣,史海蛟,等.以“痰瘀毒”立论探讨大柴胡汤防治动脉粥样硬化的炎症反应[J].辽宁中医药大学学报,2020,22(8):94-96. [22] 郑娴,王凤荣.大柴胡汤对动脉粥样硬化家兔血脂及炎性因子表达的影响[J].中医杂志,2013,54(19):1681-1685. [23] 温尔刚,高杰,丁怡铭,等.巨噬细胞MED1缺失促进雌性ApoE和LDLR敲除小鼠动脉粥样硬化发展[J].西安交通大学学报(医学版),2023,44(1):89-94. [24] 曹盼夏,彭紫凝,刘珊珊,等.腺苷酸活化蛋白激酶介导巨噬细胞脂肪酸氧化:中药防治动脉粥样硬化的途径[J].中国组织工程研究,2025,29(18):3906-3914. [25] 郭泽浩.家族性高甘油三酯血症外泌体lncRNA表达特点及调控通路探究[D].北京:北京协和医学院,2020. [26] 阚昊.高脂肪饮食对小鼠主动脉细胞组成和功能的影响及机制研究[D].无锡:江南大学,2023. [27] 杨珺,殷鹏,郑中华.水苏碱激活自噬改善高脂喂养小鼠动脉粥样硬化的作用机制[J].中国组织工程研究,2025,29(24):5140-5147. [28] 杨牧祥,马全庆,田元祥,等.脂调康胶囊对高脂血症大鼠血清IL-6的影响[J].中国中医基础医学杂志,2003,9(9):12-13. [29] 黄汝哨.血清MMP-2及基因多态性对颈动脉斑块易损性及动脉粥样硬化性脑梗死的影响[D].福州:福建医科大学,2021. [30] 毛萍,吕方超,徐晨凯,等.冠心宁通过TNFAIP3-ASK1/JNK通路对动脉粥样硬化血管平滑肌细胞表型转换的调控作用[J].心脑血管病防治,2024, 24(10):5-9+18. [31] 刘燕,高茸,汤小涵,等.基于网络药理学和细胞实验探讨柚皮苷抗动脉粥样硬化的作用机制[J].空军航空医学,2024,41(3):236-242. [32] Azevedo Martins JM, Rabelo-Santos SH, do Amaral Westin MC, et al. Tumoral and stromal expression of MMP-2,MMP-9,MMP-14,TIMP-1,TIMP-2,and VEGF-A in cervical cancer patient survival:a competing risk analysis. BMC Cancer. 2020;20(1):660. [33] 李洪涛.环氧合酶与动脉粥样硬化[J].心血管病学进展,2004,25(1):62-65. [34] 姚明鹤,闫海峰,王静,等.基于网络药理学探讨荷丹片治疗高脂血症的作用机制[J].天津中医药大学学报,2021,40(6):783-789. [35] 吴佩洋.Apoe~(-/-)和Ldlr~(-/-)小鼠动脉粥样硬化发生发展及造血系统变化的对比研究[D].上海:上海师范大学,2024. [36] 曾静,花雷,阳勇,等.异功散通过调控脑水液代谢改善APP/PS1转基因小鼠的学习记忆能力[J].南方医科大学学报,2024,44(10):2015-2023. |
| [1] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [2] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [3] | Zhou Panpan, Cui Yinglin, Zhang Wentao, Wang Shurui, Chen Jiahui, Yang Tong . Role of cellular autophagy in cerebral ischemic injury and the regulatory mechanism of traditional Chinese medicine [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1650-1658. |
| [4] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [5] | Chen Yueping, Chen Feng, Peng Qinglin, Chen Huiyi, Dong Panfeng . Based on UHPLC-QE-MS, network pharmacology, and molecular dynamics simulation to explore the mechanism of Panax notoginseng in treating osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1751-1760. |
| [6] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [7] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [8] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [9] | Liu Lingyun, He Guixin, Qin Weibin, Song Hui, Zhang Liwen, Tang Weizhi, Yang Feifei, Zhu Ziyi, Ou Yangbin . Improvement of myocardial injury by traditional Chinese medicine: mitochondrial calcium homeostasis mediates macrophage autophagy and pyroptosis pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1276-1284. |
| [10] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [11] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [12] | Fang Yuan, Qian Zhiyong, He Yuanhada, Wang Haiyan, Sha Lirong, Li Xiaohe, Liu Jing, He Yachao, Zhang Kai, Temribagen. Mechanism of Mongolian medicine Echinops sphaerocephalus L. in proliferation and angiogenesis of vascular endothelial cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7519-7528. |
| [13] | Chen Ying, Guo Xiaojing, Mo Xueni, Ma Wei, Wu Shangzhi, Li Xiangling, Xie Tingting. Analysis of diagnostic biomarkers for ischemic stroke and experimental validation of targeted cuproptosis related genes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7562-7570. |
| [14] | Zhu Chuanxi, Qiu Long, Li Lingxu, Ji Guangcheng. Chinese herbal prescription combined with head acupuncture exercise therapy improves limb spasticity in rats with ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7571-7577. |
| [15] | Zhang Yixuan, Li Dongna, Liu Chunyan. Pathological processes, inflammatory responses, and related biomarkers of periodontitis: a multi-omics analysis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7601-7610. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||