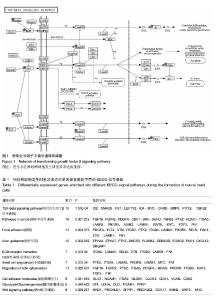
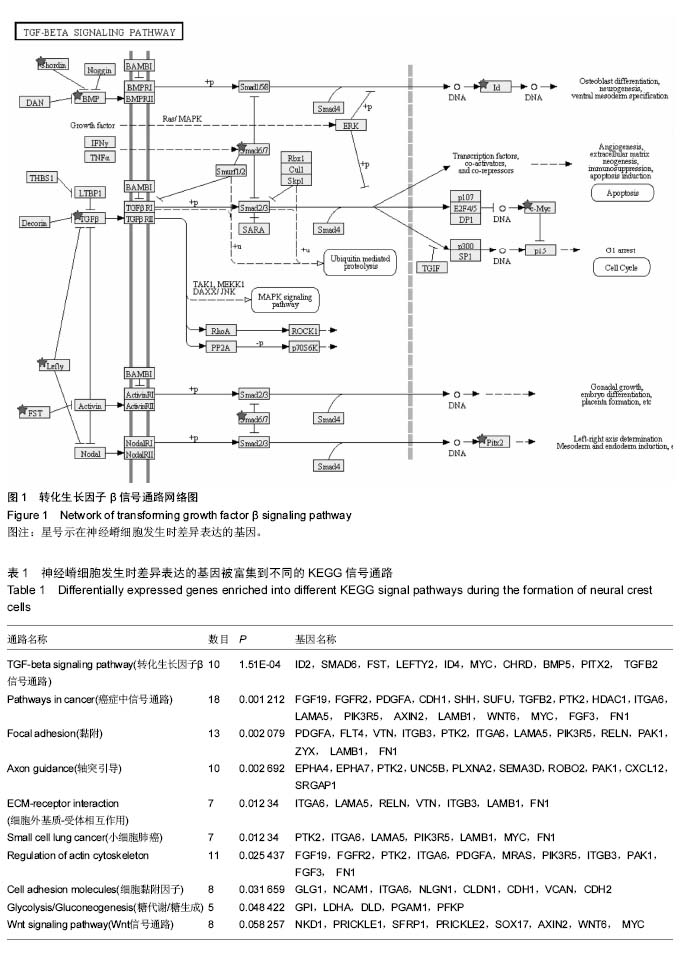
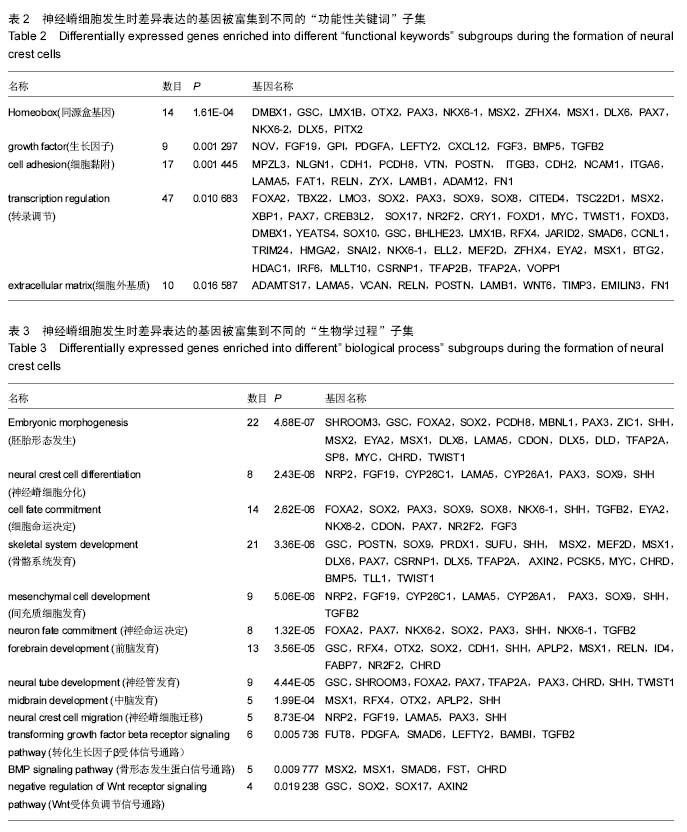
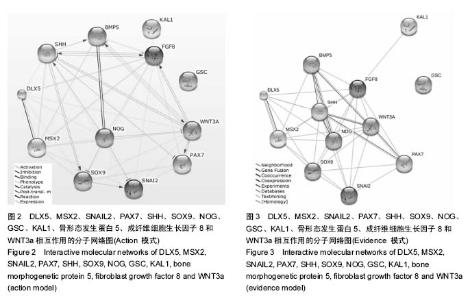
| [1] 卢境婷,王旭东,代杰文,等.颅神经嵴细胞的迁移及特性[J].中华口腔医学研究杂志:电子版,2011,5(6):58-61.
[2] Mishina Y, Snider TN. Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res. 2014;325(2):138-147.
[3] 吕红兵,金岩.神经嵴细胞的特性和培养[J]. 现代口腔医学杂志2004,18(2):181-84.
[4] Kanakubo S, Nomura T, Yamamura K,et al. Abnormal migration and distribution of neural crest cells in Pax6 heterozygous mutant eye, a model for human eye diseases. Genes Cells.2006;11(8):919-933.
[5] Song Z, Liu C, Iwata J, et al.Mice with Tak1 deficiency in neural crest lineage exhibit cleft palate associated with abnormal tongue development. J Biol Chem 2013; 288(15):10440-10450.
[6] Dai J, Kuang Y, Fang B,et al. The effect of overexpression of Dlx2 on the migration, proliferation and osteogenic differentiation of cranial neural crest stem cells. Biomaterials.2013;34(8):1898-1910.
[7] 赵晓晴,黄国英.心脏神经嵴发育与心脏锥干部畸形[J]. 国外医学:儿科学分册 2004;31(1):39-42.
[8] Dorsky RI,Moon RT,Raible DW.Control of neural crest cell fate by the Wnt signalling pathway. Nature.1998; 396(6709):370-373.
[9] Hosokawa R, Oka K, Yamaza T,et al.TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis.Dev Biol 2010;341(1):186-195.
[10] Lee J, Corcoran A, Han M,et al. Dlx5 and Msx2 regulate mouse anterior neural tube closure through ephrinA5-EphA7. Dev Growth Differ.2013;55(3): 341-349.
[11] Dongkyun K, Jinsoo S, Jin EJ. Wnt-3 and Wnt-3a play different region-specific roles in neural crest development in avians. Cell Biol Int.2010;34(7): 763-768.
[12] Endo Y,Ishiwata-Endo H,Yamada KM. Extracellular matrix protein anosmin promotes neural crest formation and regulates FGF, BMP, and WNT activities. Dev Cell. 2012;23(2):305-316.
[13] Huang da W, Sherman BT, Lempicki RA.Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc.2009;4(1): 44-57.
[14] von Mering C,Jensen LJ,Snel B, et al.STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433-437.
[15] Zhang ZG, Cao H, Liu G,et al. Bioinformatic analysis of microarray data reveals several key genes related to heart failure. Eur Rev Med Pharmacol Sci. 2013;17(18): 2441-2448. |