Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (11): 2712-2726.doi: 10.12307/2026.119
Previous Articles Next Articles
Neuroprotective regulation of the IRF9 gene after spinal cord injury: bioinformatics analysis combined with experimental validation
Tian Minghao, Liao Yehui, Zhou Wenyang, He Baoqiang, Leng Yebo, Xu Shicai, Zhou Jiajun, Li Yang, Tang Chao, Tang Qiang, Zhong Dejun
- Department of Orthopedics, Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan Province, China
-
Received:2025-02-07Accepted:2025-06-19Online:2026-04-18Published:2025-09-02 -
Contact:Zhong Dejun, Professor, Master’s supervisor, Department of Orthopedics, Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan Province, China -
About author:Tian Minghao, MS candidate, Department of Orthopedics, Affiliated Hospital of Southwest Medical University, Luzhou 646000, Sichuan Province, China -
Supported by:Sichuan Medical Association Project, No. Q22008 (to LYH); Luzhou Municipal People’s Government and Southwest Medical University Science and Technology Strategic Cooperation Project, No. 2023LZXNYDJ038 (to LYH); Doctoral Research Initiation Fund Project of the Affiliated Hospital of Southwest Medical University, No. 22155 (to LYH); Applied Basic Research Program of Southwest Medical University, No. 2023QN070 (to LYH); Natural Science Foundation of Sichuan Province, No. 2024NSFSC0682 (to ZDJ); Southwest Medical University Scientific Research Fund Program, No. 2024LCYXZX38 (to ZDJ)
CLC Number:
Cite this article
Tian Minghao, Liao Yehui, Zhou Wenyang, He Baoqiang, Leng Yebo, Xu Shicai, Zhou Jiajun, Li Yang, Tang Chao, Tang Qiang, Zhong Dejun . Neuroprotective regulation of the IRF9 gene after spinal cord injury: bioinformatics analysis combined with experimental validation[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(11): 2712-2726.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
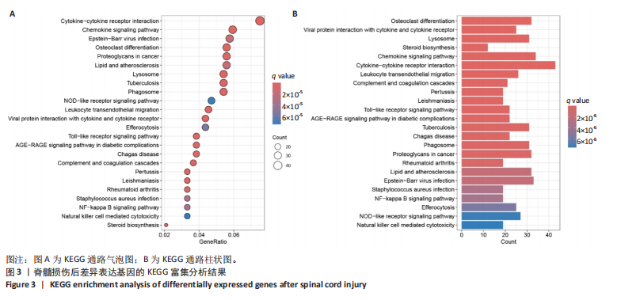
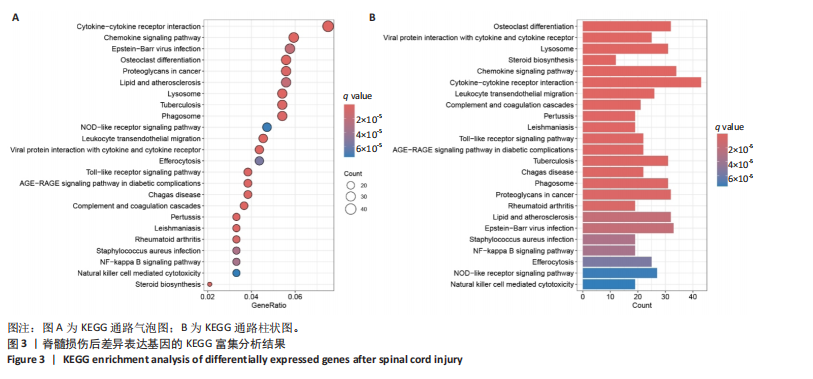
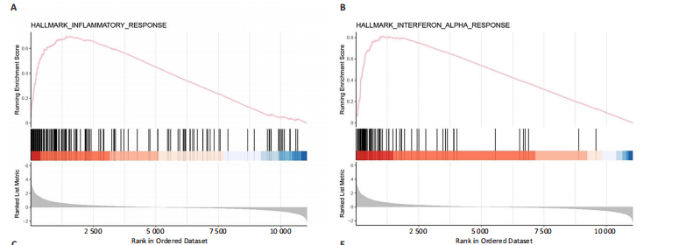
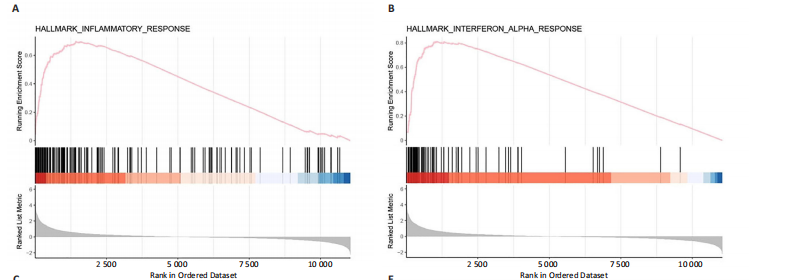
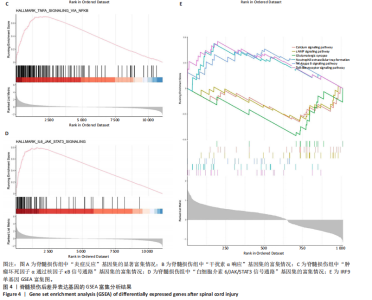
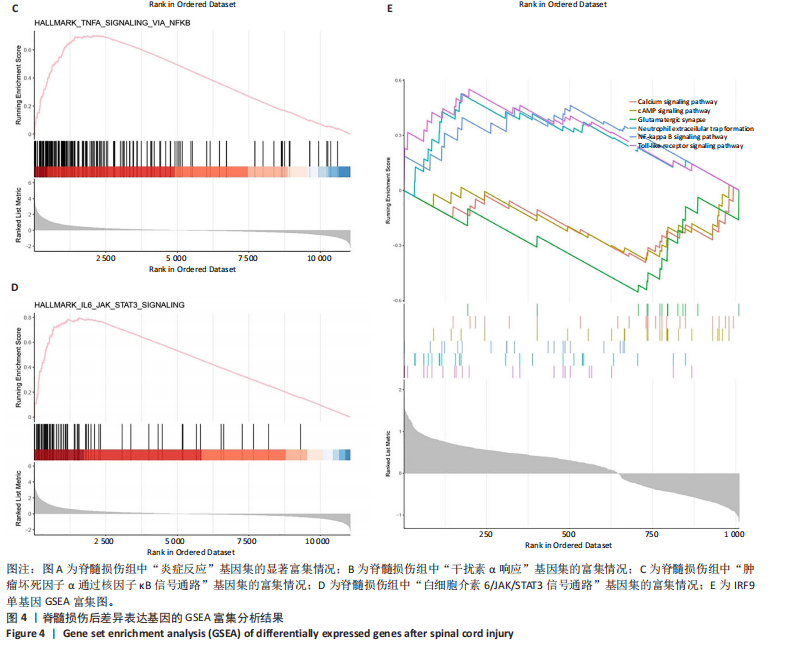
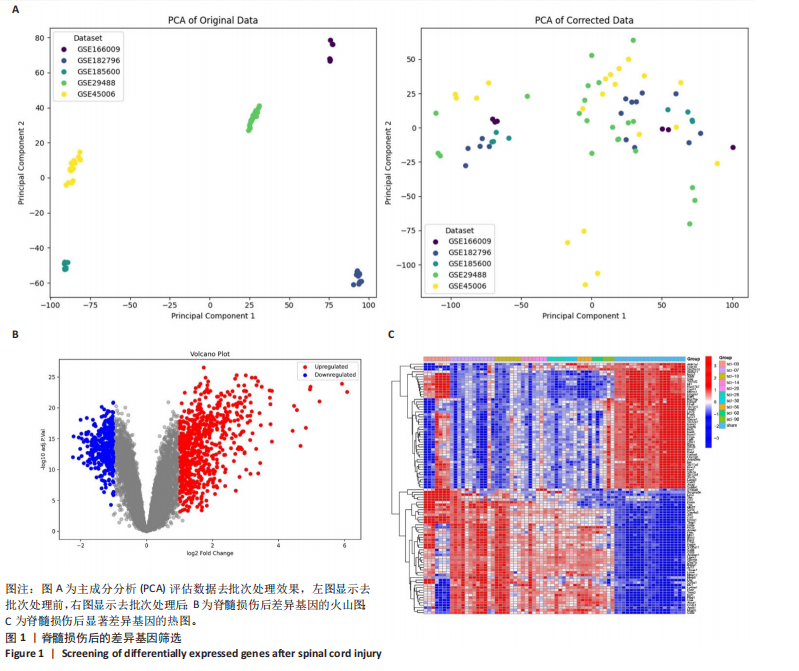
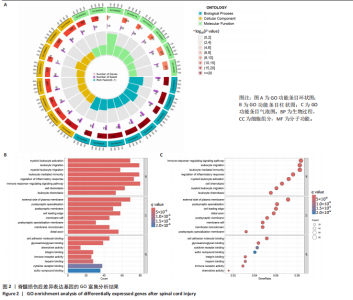
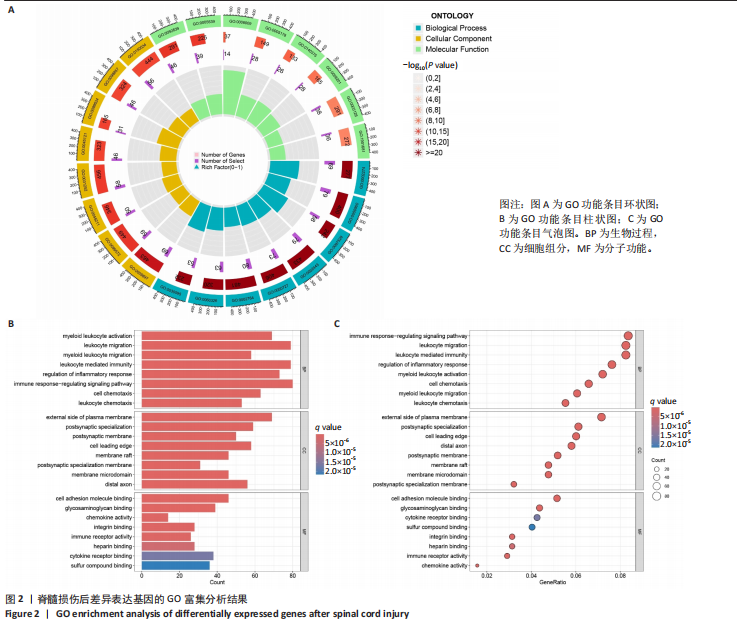
2.1 脊髓损伤后的差异基因 为探讨脊髓损伤后的基因表达变化,此次研究对多数据集进行整合分析。主成分分析结果显示,在去批次处理前,不同数据集的样本在主成分空间中呈现明显分离,表明存在显著的批次效应(图1A左)。经过ComBat方法去批次处理后,样本间的批次效应显著减少,损伤组与对照组在主成分空间中的聚类更加紧密(图1A右),验证了去批次处理的有效性,为后续的差异基因分析奠定了基础。 基于去批次处理后的数据,选择脊髓损伤后3 d以上的样本为实验组,假手术组和空白组样本为对照组,通过差异基因分析共筛选出显著差异表达基因1 041个,其中上调基因699个,下调基因342个(图1B),该火山图清晰地展示了差异基因的分布:红色点表示上调基因,蓝色点表示下调基因,灰色点为无显著差异的基因。进一步分析绘制热图(图1C),展示了前100个显著上调和下调基因在损伤组与对照组中的表达趋势,上调基因主要涉及免疫调控和炎症反应激活(例如S100a9、Serpina3n和Lcn2),其中一些基因与炎症相关过程密切相关(例如Ccl2、Timp1和Cd68);此外,还发现涉及组织修复和细胞增殖的基因显著上调,例如Mki67和Postn,下调基因主要与神经功能和神经保护相关,例如 Kcnc1和Slc17a6,这些基因涉及突触传递、神经修复及神经元功能的调控(例如 Nefh和Stx1b)。这些结果表明,脊髓损伤可能通过显著激活炎症反应并抑制神经保护机制,导致神经功能障碍的发生。 2.2 GO和KEGG富集分析结果 GO功能富集分析(图2)显示,差异基因显著富集于免疫反应、细胞迁移、神经修复及细胞微环境调控相关的功能条目。在生物过程中,富集条目如“白细胞趋化性”“免疫反应调节”和“炎症反应调节”提示脊髓损伤后免疫系统和炎症反应的显著激活。在细胞组分中,“髓样白细胞迁移”和“髓样白细胞激活”进一步表明免疫细胞在损伤部位的动员和参与,“突触后膜”“远端轴突”和“细胞前缘”暗示突触结构重塑及细胞迁移对修复的关键作用。在分子功能中,“糖胺聚糖结合”和“整合素结合”等条目表明炎症调节、细胞信号传导以及免疫细胞招募的显著激活。 KEGG通路富集分析(图3)表明,差异基因集中于“细胞因子-受体相互作用”“趋化因子信号通路”和“Toll样受体信号通路”等免疫和炎症相关通路,突出免疫细胞招募及炎症调节的重要性。“核因子kB信号通路”和“补体与凝血级联反应”提示局部炎症信号的放大过程。“白细胞经内皮迁移”和“溶酶体”通路进一步揭示免疫细胞迁移与清除的关键意义,同时支持其在神经损伤修复中的潜在功能。 2.3 GSEA富集分析结果 GSEA分析结果显示,脊髓损伤后炎症反应和免疫调控相关基因集显著富集(图4)。在全局分析中,“干扰素γ响应”(NES=2.571,P.adjust=5.41×10-26)和“干扰素α响应”(NES=2.459,P.adjust=6.26 × 10-16)在脊髓损伤组中富集程度最高,提示干扰素信号在炎症反应和免疫调控中的核心地"
位;此外,“白细胞介素6-JAK-STAT3信号通路”(NES= 2.373,P.adjust=3.08×10-12)和“肿瘤坏死因子信号通路”(NES=2.324,P.adjust=3.71×10-17)等通路也表现出显著富集,进一步支持了脊髓损伤后免疫系统激活和炎症调控的重要机制;“炎症反应”基因集的显著富集(NES=2.283,P.adjust=7.96×10-16)与GO和KEGG富集分析结果高度一致,表明炎症信号通路是脊髓损伤后病理机制的重要组成部分。 结合单基因GSEA分析,进一步探讨了IRF9在脊髓损伤中的潜在作用。结果显示,IRF9上调的通路集中在炎症和免疫反应中,其中 Toll样受体信号通路(NES=2.018,P.adjust=0.000 6)和核因子κB信号通路(NES=1.633,P.adjust=0.039 3)显著富集,提示这些通路可能通过放大局部炎症和驱动免疫细胞活化在脊髓损伤继发性损伤中发挥重要作用。另外,IRF9与中性粒细胞胞外诱捕网形成(NES=1.950,P.adjust=0.002 3)的相关性进一步说明IRF9可能通过促进急性炎症反应,加剧继发性损伤。与此同时,多个神经相关通路显著下调,其中环磷酸腺苷信号通路(NES=-1.674,P.adjust=0.025 2)和钙信号通路(NES=-1.716,P.adjust=0.014 8)显著富集,这些通路对神经元信号传递和功能修复至关重要。谷氨酸能突触(NES=-2.057 684 526,P.adjust=0.002 1)的下调可能进一步加剧了神经功能障碍和退行性病变。 综合来看,IRF9在脊髓损伤后通过激活炎症和抑制神经信号传导双重作用,可能是继发性损伤的重要调控因子,为炎症干预和神经修复研究提供了潜在靶点。"
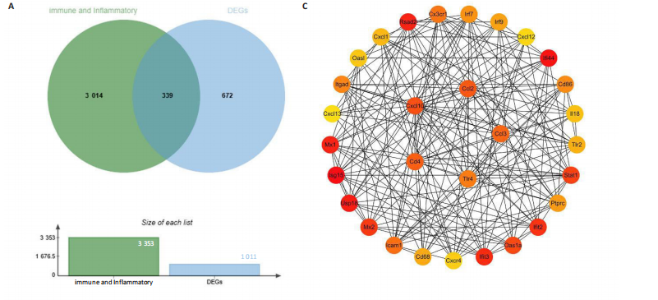
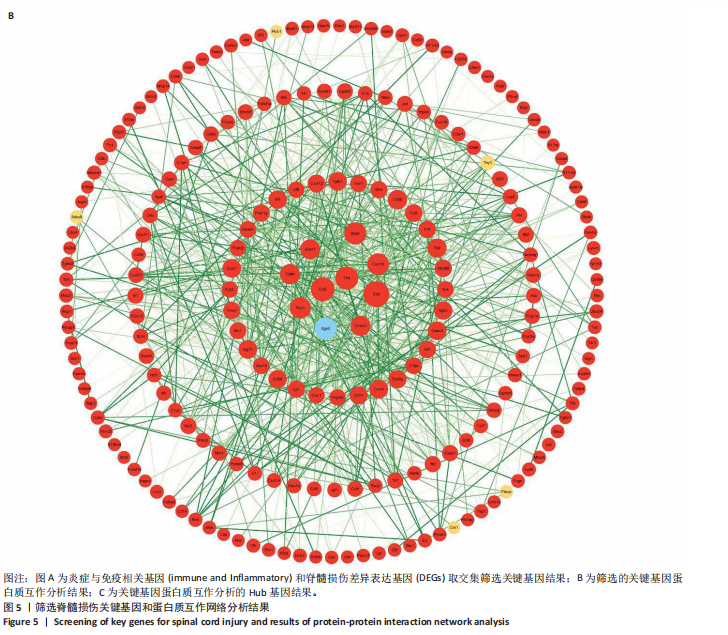
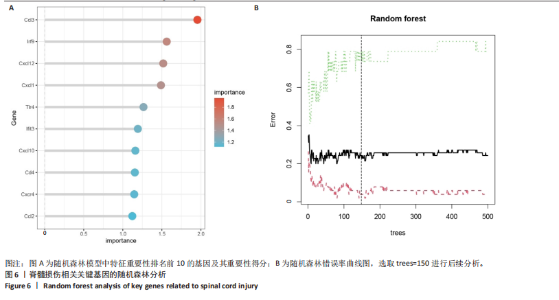
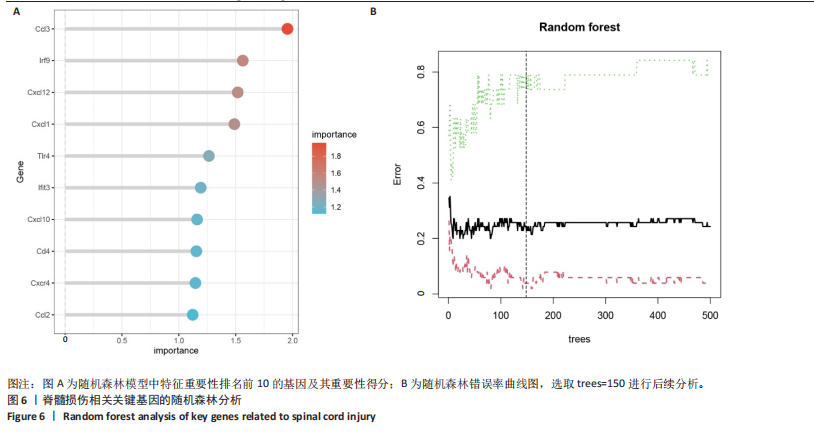
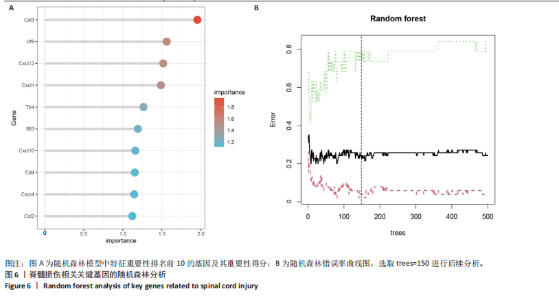
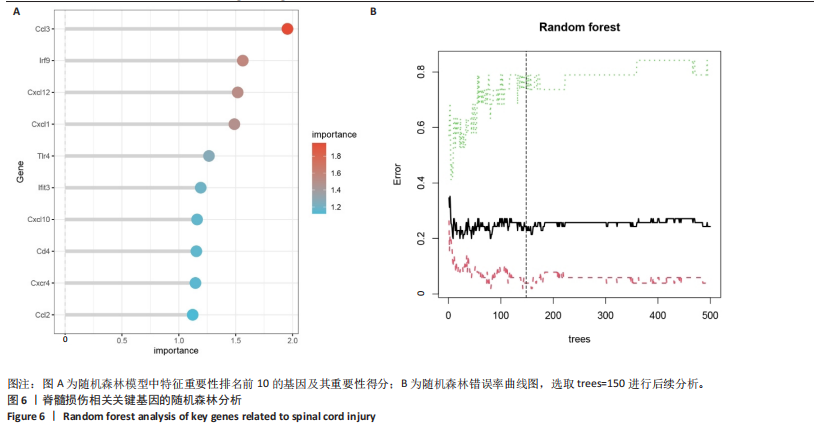
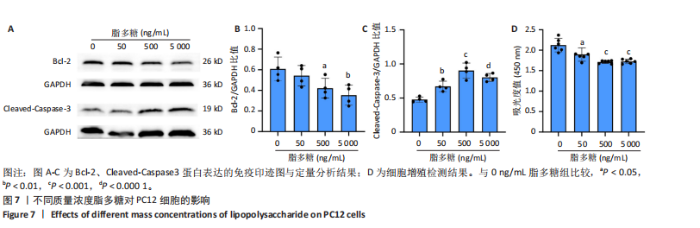
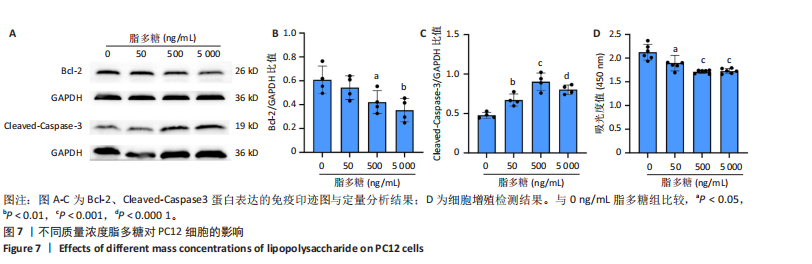
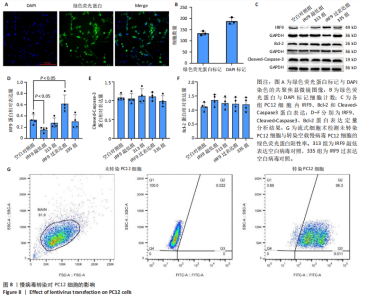
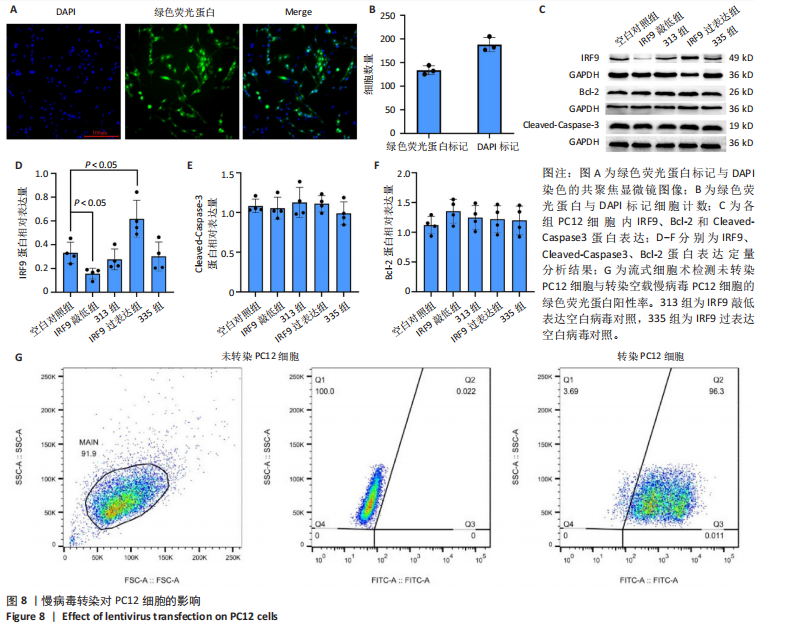
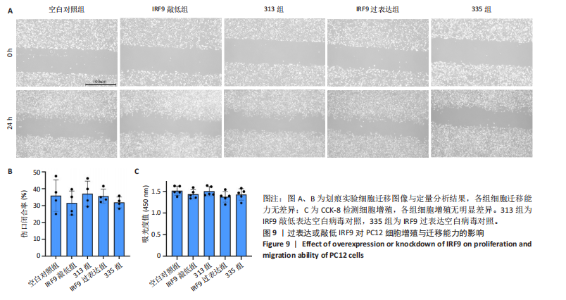
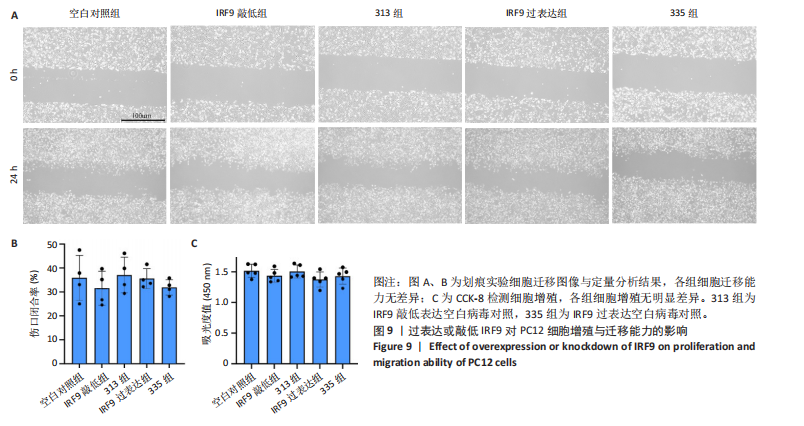
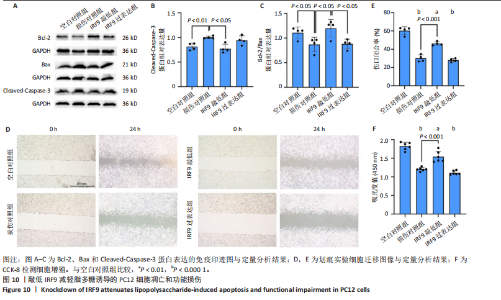
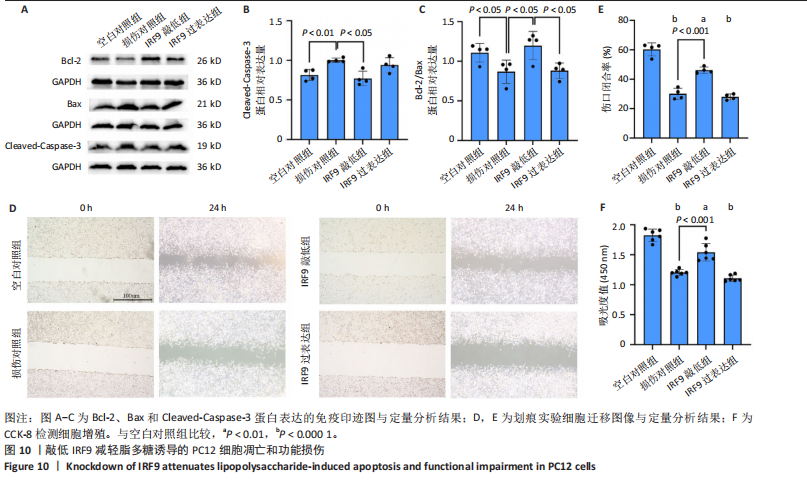
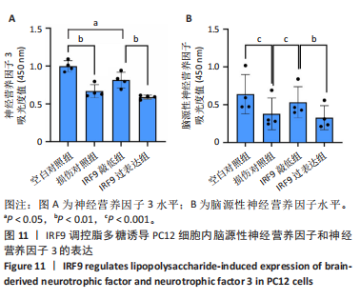
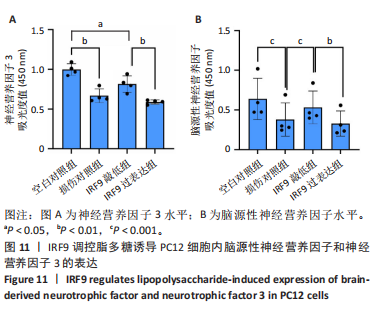
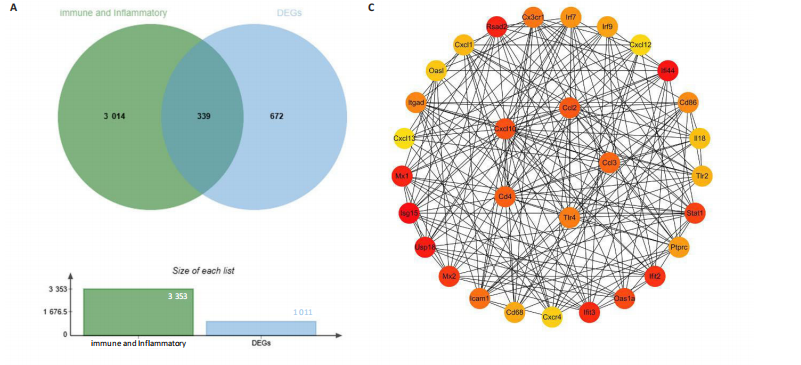
2.4 关键基因的筛选 为进一步探讨脊髓损伤过程中炎症和免疫调控的分子机制,此次研究结合前文差异基因分析结果,聚焦于与炎症和免疫相关基因集。在GO数据库中筛选基因过程如下:以“大鼠”为物种背景,分别使用关键词“inflammatory” 和 “immune” 进行检索,得到与炎症相关的基因共计 841个;免疫相关的基因共计3 115个。将两部分基因合并后,去重得到炎症和免疫相关基因总计3 353个。最后通过与差异基因交集筛选出目标基因(图5A)。这些基因既具有显著差异表达,又在免疫和炎症信号通路中发挥潜在作用,最终筛选出339个候选基因。这些基因作为后续随机森林分析和蛋白质互作网络构建的研究对象。 2.5 蛋白质互作网络分析结果 构建的蛋白质互作网络由208个节点和957条边组成,展示了差异基因之间复杂的相互作用关系(图5B)。在网络中,度值最高的5个基因分别是Cd4、Ccl2、Tlr4、Stat1和Itgad,这些基因在免疫调控和炎症反应中起着核心作用,而IRF9处于网络的次核心位置。进一步分析显示,30个Hub 基因(图5C)中同样包括IRF9,这进一步支持IRF9在脊髓损伤后的潜在功能性地位。推测IRF9 通过调控干扰素信号通路及其下游炎症因子的表达,可能在脊髓损伤后的炎症反应中发挥关键作用,调节免疫反应的平衡,进而影响组织修复和损伤进程。 2.6 随机森林分析结果 使用随机森林分析蛋白互作网络分析计算出的前30个Hub基因,筛选中重要性排名前10的Hub基因(图6),分别是:Ccl3、IRF9、Cxcl12、Cxcl1、Tlr4、Ifit3、Cxcl10、Cd4、Cxcr4、Ccl2。其中,Ccl3、Ccl2、Cxcl12、Cxcl10和 Cxcl1 作为关键趋化因子,主要参与免疫细胞募集和炎症放大效应,属于下游执行者;Tlr4是模式识别受体,位于免疫和炎症反应的最上游,通过感知损伤信号触发下游炎症级联;IRF9作为干扰素信号通路的核心转录因子,广泛调控多条炎症和免疫相关通路,是网络中重要的枢纽基因。这些基因的功能分工揭示了脊髓损伤后炎症和免疫调控的分层机制,并为探索潜在的治疗靶点提供了依据。 2.7 体外细胞实验验证结果 2.7.1 确定最适脂多糖质量浓度 Western blot检测结果显示(图7A-C),经脂多糖处理后,PC12细胞内Bcl-2蛋白表达降低、Cleaved-Caspase3蛋白表达升高,表明细胞凋亡显著增强,其中以500,5 000 ng/mL脂多糖的处理结果更明显。CCK-8细胞检测结果显示,经脂多糖处理后,PC12细胞增殖显著降低,其中以500,5 000 ng/mL脂多糖的处理结果更明显(图7D)。综合上述结果,500 ng/mL被确定为脂多糖诱导PC12细胞炎症反应和凋亡的最适质量浓度。 2.7.2 慢病毒转染率及对PC12细胞的影响 流式细胞术、DAPI染色和Western blot检测结果表明,慢病毒成功转染PC12细胞,转染效率满足后续实验需求(图8)。其中,流式细胞术和DAPI染色显示,绿色荧光蛋白阳性细胞占总细胞数量的比例超过70%;Western blot检测结果表明,5组PC12细胞内Cleaved-Caspase3、Bcl-2蛋白表达比较差异无显著性意义(P > 0.05)。划痕实验结果显示,5组PC12细胞迁移能力比较差异无显著性意义(P > 0.05),见图9A,B。CCK-8检测结果显示,5组PC12细胞增殖比较差异无显著性意义(P > 0.05),见图9C。结果表明,慢病毒转染后不会对PC12细胞造成损伤或诱导细胞凋亡。 2.7.3 敲低IRF9减轻脂多糖诱导的PC12细胞凋亡和功能损伤 Western blot检测结果显示,与空白对照组相比,损伤对照组Bax和Cleaved-Caspase3蛋白表达升高(P < 0.01或P < 0.05),Bcl-2蛋白表达降低(P < 0.05);与损伤对照组比较,IRF9敲低组Bax和Cleaved-Caspase3蛋白表达降低(P < 0.05),Bcl-2蛋白表达升高(P < 0.05),IRF9过表达组Bax、Bcl-2和Cleaved-Caspase3蛋白表达与损伤对照组比较差异无显著性意义(P > 0.05),见图10A-C。表明IRF9在脂多糖诱导的PC12细胞凋亡过程中起促进作用,敲低IRF9能够减轻细胞凋亡,但尚不能证明过表达IRF9能促进炎症刺激后的细胞凋亡。 划痕实验结果显示,与空白对照组比较,损伤对照组、IRF9过表达组、IRF9敲低组PC12细胞迁移能力均降低;与损伤对照组比较,IRF9敲低组PC12细胞迁移能力升高,见图10D,E。CCK-8检测结果显示,与空白对照组比较,损伤对照组、IRF9过表达组、IRF9敲低组PC12细胞增殖均降低;与损伤对照组比较,IRF9敲低组PC12细胞增殖升高,见图10F。 2.7.4 IRF9调控炎症损伤后PC12细胞中脑源性神经营养因子和神经营养因子3的表达 Elisa检测结果显示,与空白对照组比较,损伤对照组、IRF9敲低组、IRF9过表达组神经营养因子3水平均降低,损伤对照组、IRF9过表达组脑源性神经营养因子水平降低;与损伤对照组比较,IRF9敲低组脑源性神经营养因子水平升高,见图11。"
| [1] GBD Spinal Cord Injuries Collaborators. Global, regional, and national burden of spinal cord injury, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol. 2023;22(11):1026-1047. [2] AHUJA CS, WILSON JR, NORI S, et al. Traumatic spinal cord injury. Nat Rev Dis Primers. 2017;3:17018. [3] ALIZADEH A, DYCK SM, KARIMI-ABDOLREZAEE S. Traumatic Spinal Cord Injury: An Overview of Pathophysiology, Models and Acute Injury Mechanisms. Front Neurol. 2019;10:282. [4] 汪晶,李伦兰,丁杨,等.脊髓损伤患者住院医疗费用及其影响因素分析[J].中国病案,2021,22(4):50-54. [5] 王超宇,亢毅,娄永富,等.多中心创伤性颈脊髓损伤流行病学分析[J].中国脊柱脊髓杂志,2023,33(5):408-416. [6] ALLISON DJ, DITOR DS. Immune dysfunction and chronic inflammation following spinal cord injury. Spinal Cord. 2015;53(1): 14-18. [7] MILICH LM, RYAN CB, LEE JK. The origin, fate, and contribution of macrophages to spinal cord injury pathology. Acta Neuropathol. 2019;137(5):785-797. [8] ORR MB, GENSEL JC. Spinal Cord Injury Scarring and Inflammation: Therapies Targeting Glial and Inflammatory Responses. Neurotherapeutics. 2018;15(3):541-553. [9] SHECHTER R, SCHWARTZ M. Harnessing monocyte-derived macrophages to control central nervous system pathologies: no longer ‘if’ but ‘how’. J Pathol. 2013;229(2):332-346. [10] 樊保佑,冯世庆,陈琳,等.脊髓损伤神经修复临床治疗指南(IANR/CANR 2019年版)[J].西部医学,2020,32(6):790-802. [11] HORVATH CM, STARK GR, KERR IM, et al. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16(12):6957-6964. [12] MARTINEZ-MOCZYGEMBA M, GUTCH MJ, FRENCH DL, et al. Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem. 1997;272(32):20070-20076. [13] VEALS SA, SCHINDLER C, LEONARD D, et al. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12(8):3315-3324. [14] PAUL A, TANG TH, NG SK. Interferon Regulatory Factor 9 Structure and Regulation. Front Immunol. 2018;9:1831. [15] RENGACHARI S, GROISS S, DEVOS JM, et al. Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function. Proc Natl Acad Sci U S A. 2018;115(4):E601-609. [16] ZENG C, ZHU X, LI H, et al. The Role of Interferon Regulatory Factors in Liver Diseases. Int J Mol Sci. 2024;25(13):6874. [17] GANTA VC, CHOI MH, KUTATELADZE A, et al. A MicroRNA93-Interferon Regulatory Factor-9-Immunoresponsive Gene-1-Itaconic Acid Pathway Modulates M2-Like Macrophage Polarization to Revascularize Ischemic Muscle. Circulation. 2017;135(24):2403-2025. [18] ZHANG Y, LIU X, SHE ZG, et al. Interferon regulatory factor 9 is an essential mediator of heart dysfunction and cell death following myocardial ischemia/reperfusion injury. Basic Res Cardiol. 2014; 109(5):434. [19] ZHANG SM, ZHU LH, CHEN HZ, et al. Interferon regulatory factor 9 is critical for neointima formation following vascular injury. Nat Commun. 2014;5:5160. [20] CHEN HZ, GUO S, LI ZZ, et al. A critical role for interferon regulatory factor 9 in cerebral ischemic stroke. J Neurosci. 2014;34(36):11897-11912. [21] LEI J, ZHOU MH, ZHANG FC, et al. Interferon regulatory factor transcript levels correlate with clinical outcomes in human glioma. Aging (Albany NY). 2021;13(8):12086-12098. [22] DAS A, CHAI JC, KIM SH, et al. Transcriptome sequencing of microglial cells stimulated with TLR3 and TLR4 ligands. BMC Genomics. 2015; 16(1):517. [23] CHAMANKHAH M, EFTEKHARPOUR E, KARIMI-ABDOLREZAEE S, et al. Genome-wide gene expression profiling of stress response in a spinal cord clip compression injury model. BMC Genomics. 2013; 14:583. [24] TURNER SMF, SUNSHINE MD, CHANDRAN V, et al. Hyperbaric Oxygen Therapy after Mid-Cervical Spinal Contusion Injury. J Neurotrauma. 2022;39(9-10):715-723. [25] LI X, YANG Z, ZHANG A. The effect of neurotrophin-3/chitosan carriers on the proliferation and differentiation of neural stem cells. Biomaterials. 2009;30(28):4978-4985. [26] JOHNSON WE, LI C, RABINOVIC A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8(1):118-127. [27] PEDREGOSA F, VAROQUAUX G, GRAMFORT A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825-2830. [28] SMYTH GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [29] HUNTER JD. Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 2007;9(3):90-95. [30] ASHBURNER M, BALL CA, BLAKE JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-29. [31] OGATA H, GOTO S, SATO K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29-34. [32] YU G, WANG LG, HAN Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5): 284-287. [33] SUBRAMANIAN A, TAMAYO P, MOOTHA VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A.2005;102(43): 15545-15550. [34] LIBERZON A, SUBRAMANIAN A, PINCHBACK R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739-1740. [35] HÄNZELMANN S, CASTELO R, GUINNEY J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [36] SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. [37] SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. [38] CHIN CH, CHEN SH, WU HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11. [39] BREIMAN L. Random Forests. Machine Learning. 2001;45(1):5-32. [40] LIAW A, WIENER MC. Classification and Regression by randomForest. R News. 2002;2(3):18-22. [41] SCHNEIDER CA, RASBAND WS, ELICEIRI KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671-675. [42] 戴静雯,周萍萍,李素,等.抗病毒免疫中干扰素与炎症信号通路的交互调控:防御反应与维持稳态 [J].微生物学报,2022, 62(10):3709-3721. [43] THIBAULT DL, CHU AD, GRAHAM KL, et al. IRF9 and STAT1 are required for IgG autoantibody production and B cell expression of TLR7 in mice. J Clin Invest. 2008;118(4):1417-1426. [44] 单佳铃,程虹毓,文乐,等.TLR/MyD 88/NF-κB信号通路参与不同疾病作用机制研究进展[J].中国药理学通报,2019,35(4):451-455. [45] MEZZASOMA L, ANTOGNELLI C, TALESA VN. A Novel Role for Brain Natriuretic Peptide: Inhibition of IL-1β Secretion via Downregulation of NF-kB/Erk 1/2 and NALP3/ASC/Caspase-1 Activation in Human THP-1 Monocyte. Mediators Inflamm. 2017;2017:5858315. [46] ZAMBRANO-ZARAGOZA JF, GUTIÉRREZ-FRANCO J, DURÁN-AVELAR MJ, et al. Neutrophil extracellular traps and inflammatory response: Implications for the immunopathogenesis of ankylosing spondylitis. Int J Rheum Dis. 2021;24(3):426-433. [47] TAO X, FINKBEINER S, ARNOLD DB, et al. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20(4):709-726. [48] LI X, WANG G, LI W, et al. Histone deacetylase 9 plays a role in sevoflurane-induced neuronal differentiation inhibition by inactivating cAMP-response element binding protein transcription and inhibiting the expression of neurotrophin-3. FASEB J. 2023;37(10):e23164. [49] ARUHAN, GONG Q, TUO YJ, et al. Syringa oblata Lindl Extract Alleviated Corticosterone-Induced Depression via the cAMP/PKA-CREB-BDNF Pathway. J Ethnopharmacol. 2025;341:119274. [50] YOUNG M, BOOTH DM, SMITH D, et al. Transcriptional regulation in the absence of inositol trisphosphate receptor calcium signaling. Front Cell Dev Biol. 2024;12:1473210. [51] NASSRALLAH WB, CHENG J, MACKAY JP, et al. Mechanisms of synapse-to-nucleus calcium signalling in striatal neurons and impairments in Huntington’s disease. J Neurochem. 2024;168(9):2671-2689. [52] LIANG CW, CHEN TC. Anti-NMDA receptor encephalitis presenting as fever with undetermined cause. Int J Infect Dis. 2022;122:365-367. [53] CHEN Q, LIANG Z, YUE Q, et al. A Neuropeptide Y/F-like Polypeptide Derived from the Transcriptome of Turbinaria peltata Suppresses LPS-Induced Astrocytic Inflammation. J Nat Prod. 2022;85(6):1569-1580. [54] HARLAND M, TORRES S, LIU J, et al. Neuronal Mitochondria Modulation of LPS-Induced Neuroinflammation. J Neurosci. 2020;40(8):1756-1765. [55] KIM J, LEE HJ, PARK JH, et al. Nilotinib modulates LPS-induced cognitive impairment and neuroinflammatory responses by regulating P38/STAT3 signaling. J Neuroinflammation. 2022;19(1):187. [56] SKRZYPCZAK-WIERCIOCH A, SAŁAT K. Lipopolysaccharide-Induced Model of Neuroinflammation: Mechanisms of Action, Research Application and Future Directions for Its Use. Molecules. 2022; 27(17):5481. [57] 吴军,丁单华,李倩倩,等.脂多糖刺激小胶质细胞激活分化的机制研究[J].中华神经医学杂志,2018,17(12):1195-1202. [58] LI C, WU Z, ZHOU L, et al. Temporal and spatial cellular and molecular pathological alterations with single-cell resolution in the adult spinal cord after injury. Signal Transduct Target Ther. 2022;7(1):65. [59] BALON K, WIATRAK B. PC12 and THP-1 Cell Lines as Neuronal and Microglia Model in Neurobiological Research. Appl Sci. 2021; 11(9):3729. [60] OPREA D, SANZ CG, BARSAN MM, et al. PC-12 Cell Line as a Neuronal Cell Model for Biosensing Applications. Biosensors (Basel). 2022;12(7):500. [61] 周涛,许百男,陈德蕙,等.PC12细胞分化的神经元与离体培养的大鼠原代皮质神经元之间功能性突触的形成[J].中华神经科杂志, 2005,38(3):183-186. [62] BIBER K, OWENS T, BODDEKE E. What is microglia neurotoxicity (Not)? Glia. 2014;62(6):841-854. [63] DAVID S, KRONER A. Repertoire of microglial and macrophage responses after spinal cord injury. Nat Rev Neurosci. 2011;12(7):388-399. [64] OLAH M, AMOR S, BROUWER N, et al. Identification of a microglia phenotype supportive of remyelination. Glia. 2012;60(2):306-321. [65] BROCKIE S, ZHOU C, FEHLINGS MG. Resident immune responses to spinal cord injury: role of astrocytes and microglia. Neural Regen Res. 2024;19(8):1678-1685. [66] PERRY VH, NICOLL JA, HOLMES C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;6(4):193-201. [67] YU H, CHANG Q, SUN T, et al. Metabolic reprogramming and polarization of microglia in Parkinson’s disease: Role of inflammasome and iron. Ageing Res Rev. 2023;90:102032. |
| [1] | Fu Lyupeng, Yu Peng, Liang Guoyan, Chang Yunbing. Electroactive materials applied in spinal surgery [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(8): 2113-2123. |
| [2] | Yuan Xiaoshuang, Yang Xu, Yang Bo, Chen Xiaoxu, Tian Ting, Wang Feiqing, Li Yanju, Liu Yang, Yang Wenxiu. Effect of conditioned medium of diffuse large B-cell lymphoma cells on proliferation and apoptosis of human bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1632-1640. |
| [3] | He Jiale, Huang Xi, Dong Hongfei, Chen Lang, Zhong Fangyu, Li Xianhui. Acellular dermal matrix combined with adipose-derived stem cell exosomes promotes burn wound healing [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1699-1710. |
| [4] | Chen Yulin, He Yingying, Hu Kai, Chen Zhifan, Nie Sha Meng Yanhui, Li Runzhen, Zhang Xiaoduo , Li Yuxi, Tang Yaoping. Effect and mechanism of exosome-like vesicles derived from Trichosanthes kirilowii Maxim. in preventing and treating atherosclerosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1768-1781. |
| [5] | Liu Anting, Lu Jiangtao, Zhang Wenjie, He Ling, Tang Zongsheng, Chen Xiaoling. Regulation of AMP-activated protein kinase by platelet lysate inhibits cadmium-induced neuronal apoptosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(7): 1800-1807. |
| [6] | Lyu Guoqing, Aizimaitijiang·Rouzi, Xiong Daohai. Irisin inhibits ferroptosis in human articular chondrocytes: roles and mechanisms [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1359-1367. |
| [7] | Peng Zhiwei, Chen Lei, Tong Lei. Luteolin promotes wound healing in diabetic mice: roles and mechanisms [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1398-1406. |
| [8] | Jia Jinwen, Airefate·Ainiwaer, Zhang Juan. Effects of EP300 on autophagy and apoptosis related to allergic rhinitis in rats [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1439-1449. |
| [9] | Yin Yongcheng, Zhao Xiangrui, Yang Zhijie, Li Zheng, Li Fang, Ning Bin. Effect and mechanism of peroxiredoxin 1 in microglial inflammation after spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1106-1113. |
| [10] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [11] | Liu Yu, Lei Senlin, Zhou Jintao, Liu Hui, Li Xianhui. Mechanisms by which aerobic and resistance exercises improve obesity-related cognitive impairment [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1171-1183. |
| [12] | Yu Huifen, Mo Licun, Cheng Leping. The position and role of 5-hydroxytryptamine in the repair of tissue injury [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1196-1206. |
| [13] | Wang Zhengye, Liu Wanlin, Zhao Zhenqun. Advance in the mechanisms underlying miRNAs in steroid-induced osteonecrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1207-1214. |
| [14] | Yang Xiao, Bai Yuehui, Zhao Tiantian, Wang Donghao, Zhao Chen, Yuan Shuo. Cartilage degeneration in temporomandibular joint osteoarthritis: mechanisms and regenerative challenges [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 926-935. |
| [15] | Bao Zhuoma, Hou Ziming, Jiang Lu, Li Weiyi, Zhang Zongxing, Liu Daozhong, Yuan Lin. Effect and mechanism by which Pterocarya hupehensis skan total flavonoids regulates the proliferation, migration and apoptosis of fibroblast-like synoviocytes [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 816-823. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||