Chinese Journal of Tissue Engineering Research ›› 2026, Vol. 30 ›› Issue (16): 4253-4264.doi: 10.12307/2026.731
Previous Articles Next Articles
Bioinformatics-based analysis of shared genes and associations in immune mechanisms between rheumatoid arthritis and Crohn’s disease
Lu Liwei1, Huang Keqi1, Chen Yueping2, Zhuo Yinghong2, Zhu Naihui1, Wei Peng1
- 1Graduate School of Guangxi University of Chinese Medicine, Nanning 530001, Guangxi Zhuang Autonomous Region, China; 2Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China
-
Received:2025-07-04Accepted:2025-08-27Online:2026-06-08Published:2025-11-29 -
Contact:Chen Yueping, PhD, Chief physician, Doctoral supervisor, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China -
About author:Lu Liwei, MS candidate, Graduate School of Guangxi University of Chinese Medicine, Nanning 530001, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, No. 81960803 (to CYP); Natural Science Foundation of Guangxi Zhuang Autonomous Region, No. 2023JJA140318 (to CYP); “Gui’s Traditional Chinese Medicine Inheritance and Innovation Team” of Guangxi University of Chinese Medicine, No. 2022A004 (to CYP)
CLC Number:
Cite this article
Lu Liwei, Huang Keqi, Chen Yueping, Zhuo Yinghong, Zhu Naihui, Wei Peng. Bioinformatics-based analysis of shared genes and associations in immune mechanisms between rheumatoid arthritis and Crohn’s disease[J]. Chinese Journal of Tissue Engineering Research, 2026, 30(16): 4253-4264.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
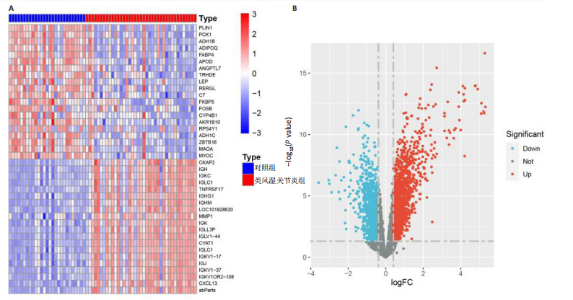
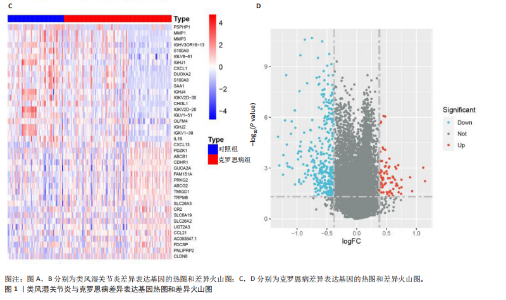
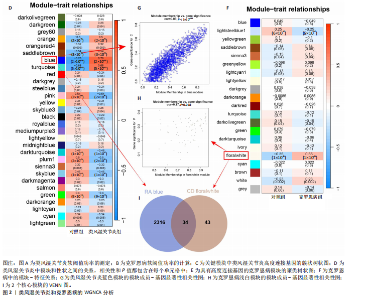
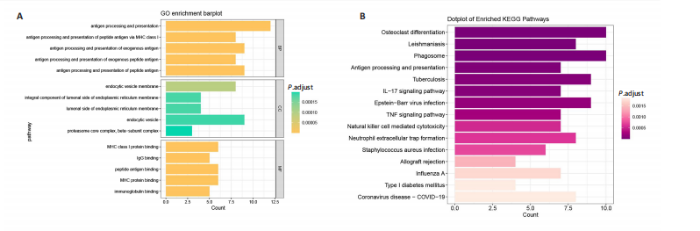
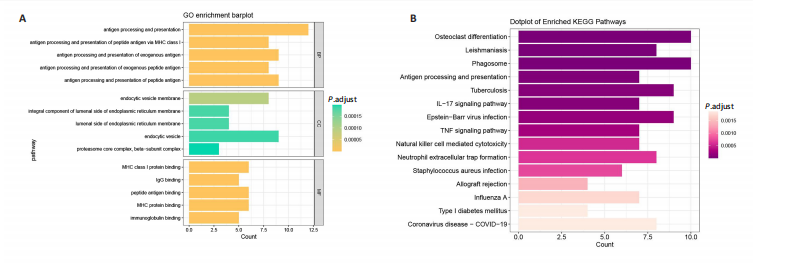
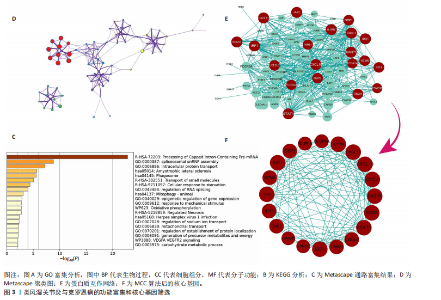
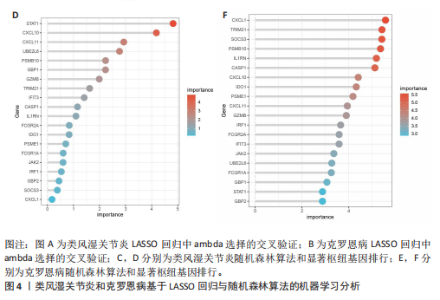
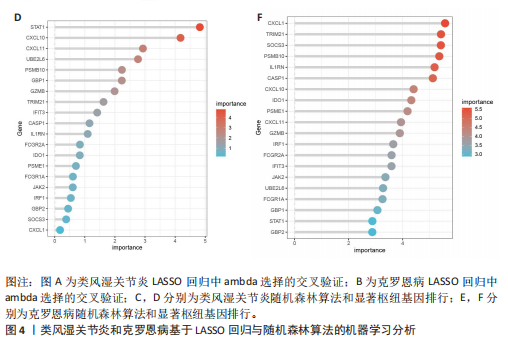
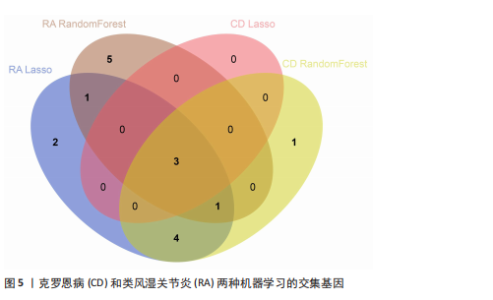
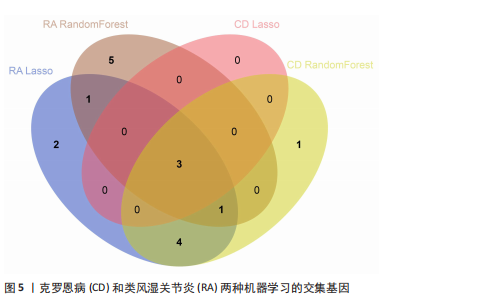
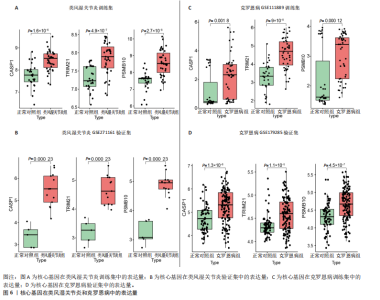
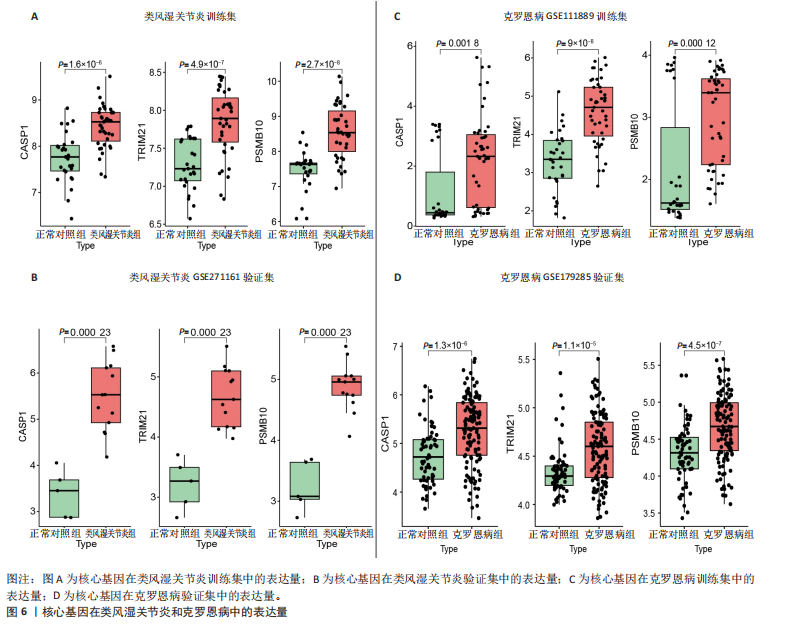
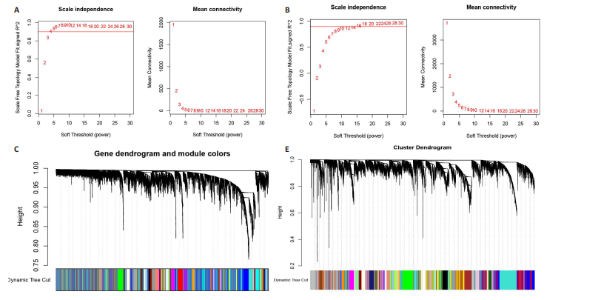
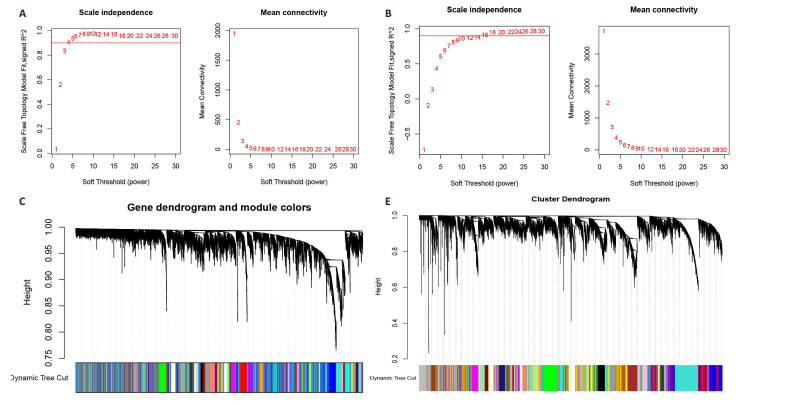
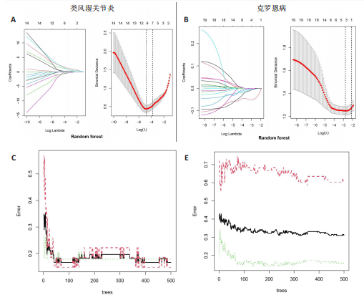
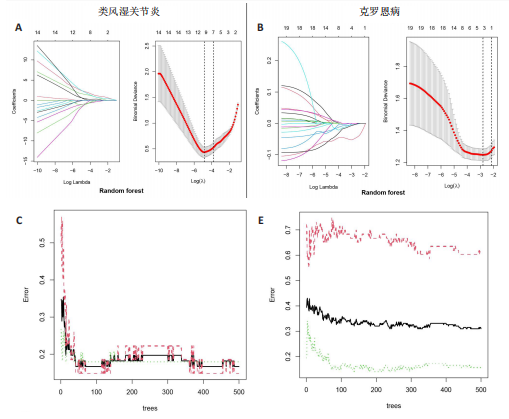
2.1 类风湿关节炎、克罗恩病和对照组的差异表达基因分析 在生物信息分析之前,先对收集的数据集进行批量效应检验,发现两种疾病的批量效应明显,使用“sva”包消除类风湿关节炎组和克罗恩病组的批量效应,然后使用limma软件包来进行分析两组之间的差异表达基因。类风湿关节炎共获得2 516个差异表达基因(adj.P < 0.05,|log2 FC| > 0.4),其中上调基因1 321个、下调基因1 195个。克罗恩病有281个差异表达基因(adj.P < 0.05,|log2 FC| > 0.38),其中上调差异40个基因、下调基因241个。火山图显示类风湿关节炎组和克罗恩病组的所有差异表达基因,两组差异最大的前40个差异表达基因通过热图展示,见图1。 2.2 WGCNA对关键模块的筛选 在类风湿关节炎数据集和克罗恩病数据集上进行WGCNA,探索临床性状与基因之间的相关性。2个数据集均未存在显著的异常样本。根据WGCNA方法,类风湿关节炎数据集中的最佳软阈值为4(图2A),克罗恩病数据集中的最佳软阈值为9(图2B)。基于模块间的相似性,最终在类风湿关节炎数据集中确定了30个模块(图2C,D),在克罗恩病数据集中确定了21个模块(图2E,F)。然后计算模块与性状之间的相关性,发现蓝色模块与类风湿关节炎的相关性最强(r=0.76),浅白色模块与克罗恩病的正相关关系最强(r=0.35)。更重要的是,模块内的基因显著性与模块成员也存在较强的正相关性(类风湿关节炎cor=0.89,克罗恩病cor=0.27),表明该模块的共表达网络结构与其生物学功能高度一致,再次证实模块基因与疾病发生显著相关(图2G,H)。最终通过WGCNA发现了34个重叠基因(图2I)。 2.3 类风湿关节炎和克罗恩病协同驱动基因的富集分析 类风湿关节炎和克罗恩病模块有34个重叠基因,差异表达基因有56个共享基因。考虑到从WGCNA筛选出的模块包含一组具有相似表达谱的基因,这些基因可能无法覆盖差异表达基因的全部范围,甚至与可能对疾病进展至关重要的差异表达基因有很大差异,因此,将差异表达基因与模块基因整合以避免遗漏。再次去除重复项后获得82个共同驱动基因,这些基因可能在类风湿关节炎和克罗恩病涉及的常见分子机制中发挥重要作用。首先对基因进行GO和KEGG富集分析(图3A,B),GO富集结果表明基因参与各种抗原的加工和呈递、内质网膜腔面和多种免疫球蛋白结合,KEGG富集结果表明基因参与破骨细胞分化、肿瘤坏死因子信号通路和白细胞介素17信号通路等通路。Metascape数据库中的不同基因可能表现出不同的功能群分布,其中含帽内含子前体mRNA的处理和RNA的代谢途径最为突出(图3C);同时Metascape数据库富集分析也显示免疫和炎症在类风湿关节炎和克罗恩病的病因学中具有共同作用(图3D)。 最后进一步筛选出同一功能组的基因,将82个候选的共同驱动基因输入到String数据库中,利用Cytoscape软件中的MCODE算法,根据上述基因及相关参数构建蛋白互作网络(图3E)。最后共发现5个基因簇,确定第一个基因簇的20个基因MCODE分均分最高,因此基因簇中的20个基因为候选核心基因(图3F),具体为IRF1、STAT1、IL1RN、CASP1、CXCL10、CXCL11、SOCS3、JAK2、CXCL1、IDO1、GBP1、UBE2L6、GZMB、TRIM21、GBP2、IFIT3、FCGR2A、FCGR1A、PSME1、PSMB10。 2.4 通过LASSO与随机森林算法识别和验证潜在的共同核心基因 为进一步筛选出最具诊断价值的关键基因,在上述20个候选基因中依次进行LASSO和随机森林模型进行筛选。采用LASSO方法筛选类风湿关节炎中的11个基因和克罗恩病中的3个基因(图4A,B)。同时,应用随机森林方法从类风湿关节炎中筛选出10个基因(图4C,D),克罗恩病保留了所有9个基因(图4E,F)。将上述不同方法在不同数据集中过滤的基因相互重叠,最终鉴定出3个常见核心基因半胱天冬酶1(CASP1)、三联基序21(TRIM21)及蛋白酶体亚基10 "

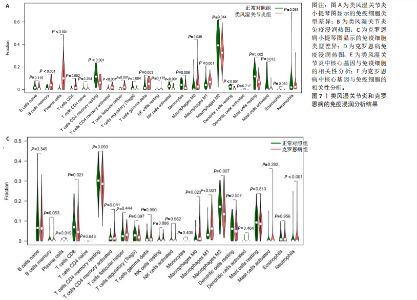
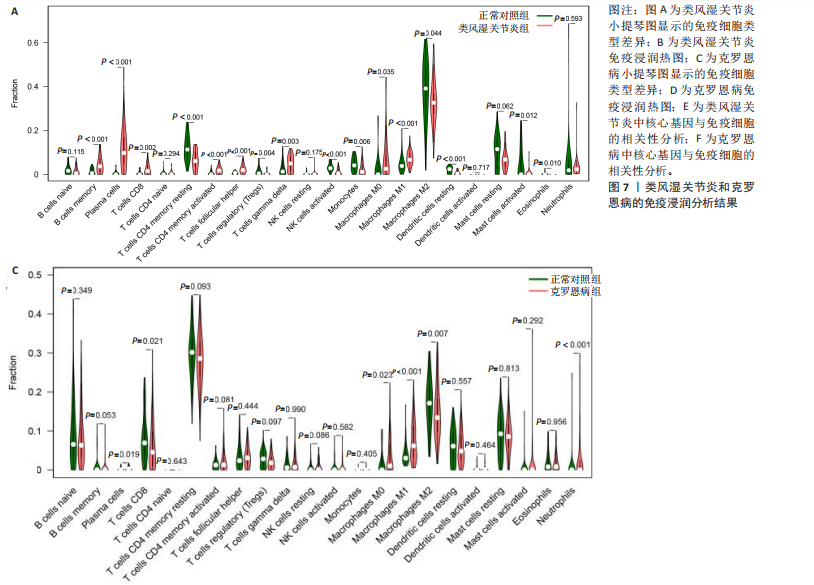
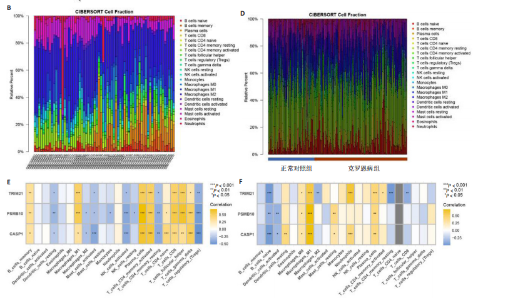
(PSMB10)(图4E)。验证集箱线图显示,类风湿关节炎和克罗恩病训练集中的所有3个诊断标志物在训练组中均显著上调(图5)。更重要的是,克罗恩病验证集和类风湿关节炎验证集都显示出一致的差异趋势(图6)。 2.5 免疫浸润分析结果 CIBERSORT方法识别22种免疫细胞不同的免疫浸润模式,首先评估类风湿关节炎数据集和克罗恩病数据集,差异表达分析结果显示,与正常对照样本相比,类风湿关节炎和克罗恩病的差异趋势一致。类风湿关节炎和克罗恩病中的巨噬细胞M0、巨噬细胞M1和中性粒细胞的表达明显更高(图7A-D),表明免疫失调和炎症反应可能发生在类风湿关节炎和克罗恩病中。然而,免疫细胞组成比例的共同差异只是类"
风湿关节炎和克罗恩病共同发病机制的一个方面,仍需确认这3个基因与病变组织的免疫浸润的关系。相关性分析显示,在类风湿关节炎训练集中,CASP1、TRIM21及PSMB10与巨噬细胞M1、浆细胞呈正相关(图7E),同时在克罗恩病训练集中也观察到类似结果(图7F),说明在克罗恩病病变组织中,巨噬细胞M1、浆细胞的浸润丰度会随着CASP1、TRIM21及PSMB10的高表达而变高。在两疾病相关性分析中,TRIM21与记忆CD4+ T细胞均呈负相关;在类风湿关节炎中,CASP1、TRIM21及PSMB10除了与记忆CD4+ T细胞和调节性T细胞呈负相关外,与T细胞多种亚型均呈正相关,这也意味着该疾病中会有更多的T细胞富集(图7E,F),再次表明核心基因可能通过调节免疫细胞的表达参与调节自身免疫。 "
| [1] CONFORTI A, DI COLA I, PAVLYCH V, et al. Beyond the joints, the extra-articular manifestations in rheumatoid arthritis. Autoimmun Rev. 2021;20(2):102735. [2] TURESSON C, MATTESON EL. Extra-articular manifestations in rheumatoid arthritis. Int J Adv Rheumatol. 2007;5(3):72. [3] RADU AF, BUNGAU SG. Management of rheumatoid arthritis: an overview. Cells. 2021;10(11):2857. [4] WU Z, MA D, YANG H,et al. Fibroblast-like synoviocytes in rheumatoid arthritis: surface markers and phenotypes. Int Immunopharmacol. 2021;93:107392. [5] RODA G, CHIEN NG S, KOTZE PG, et al. Crohn’s disease. Nat Rev Dis Primers. 2020; 6(1):22. [6] PETAGNA L, ANTONELLI A, GANINI C, et al. Pathophysiology of Crohn’s disease inflammation and recurrence. Biol Direct. 2020;15(1):23. [7] CLAYTOR J, KUMAR P, ANANTHAKRISHNAN AN, et al. Mild Crohn’s disease: definition and management. Curr Gastroenterol Rep. 2023;25(3):45-51. [8] WANG B, XIONG Y, LI R, et al. Potential role of SNP rs2071475 in rheumatoid arthritis and inflammatory bowel disease in the East Asian population: a Mendelian randomization study. Inflammopharmacology. 2024;32(1):683-692. [9] SACCON TD, DHAHBI JM, SCHNEIDER A, et al. Plasma miRNA profile of Crohn’s disease and rheumatoid arthritis patients. Biology. 2022;11(4):508. [10] ELSOURI K, ARBOLEDA V, HEISER S, et al. Microbiome in rheumatoid arthritis and celiac disease: a friend or foe. Cureus. 2021;13(6):le15543. [11] SCHER JU, ABRAMSON SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7(10):569-578. [12] WANG Q, ZHANG SX, CHANG MJ,et al. Characteristics of the gut microbiome and its relationship with peripheral CD4+ T cell subpopulations and cytokines in rheumatoid arthritis. Frontiers in Microbiology. 2022; 13:799602. [13] WELLS PM, WILLIAMS FM, MATEY-HERNANDEZ ML, et al.’RA and the microbiome: do host genetic factors provide the link? J Autoimmun. 2019;99:104-115. [14] MASSIMINO L, BARCHI A, MANDARINO FV, et al. A multi-omic analysis reveals the esophageal dysbiosis as the predominant trait of eosinophilic esophagitis. J Transl Med. 2023;21(1):46. [15] GURTNER A, CREPAZ D, ARNOLD IC. Emerging functions of tissue-resident eosinophils. J Exp Med. 2023;220(7): e20221435. [16] JACOBS I, CEULEMANS M, WAUTERS L, et al. Role of eosinophils in intestinal inflammation and fibrosis in inflammatory bowel disease: an overlooked villain? Front Immunol. 2021;12:754413. [17] FILIPPONE RT, SAHAKIAN L, APOSTOLOPOULOS V, et al. Eosinophils in inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(7):1140-1151. [18] ZOU C, BEARD JA, YANG G, et al. CASP orter: A Novel Inducible Human CASP1/NALP3/ASC Inflammasome Biosensor. J Inflamm Res. 2022;1:1183-1194. [19] INSERRA A, CHOO JM, LEWIS MD, et al. Mice lacking Casp1, Ifngr and Nos2 genes exhibit altered depressive-and anxiety-like behaviour, and gut microbiome composition. Sci Rep. 2019;9(1):6456. [20] ADDOBBATI C, DA CRUZ HL, ADELINO JE, et al. Polymorphisms and expression of inflammasome genes are associated with the development and severity of rheumatoid arthritis in Brazilian patients. Inflamm Res. 2018;67:255-264. [21] FOSS S, BOTTERMANN M, JONSSON A, et al.TRIM21—from intracellular immunity to therapy. Front Immunol. 2019;10:2049. [22] SAFONOVA TN, ZAITSEVA GV, LOGINOV VI, et al. Association of polymorphisms of the TRIM21 gene with the severity of dry keratoconjunctivitis in rheumatoid arthritis and Sjogren’s disease. Vestn Oftalmol. 2019;135(5.Vyp.2):192-198. [23] LEE AY, REED JH, GORDON TP. Anti-Ro60 and anti-Ro52/TRIM21: Two distinct autoantibodies in systemic autoimmune diseases. J Autoimmun. 2021;124:102724. [24] LIU J, ZHANG C, XU D, et al. The ubiquitin ligase TRIM21 regulates mutant p53 accumulation and gain of function in cancer. J Clin Invest. 2023;133(6):e164354. [25] LIU YX, WAN S, YANG XQ, et al. TRIM21 is a druggable target for the treatment of metastatic colorectal cancer through ubiquitination and activation of MST2. Cell Chem Biol. 2023;30(7):709-725. [26] SARRABAY G, MÉCHIN D, SALHI A, et al. PSMB10, the last immunoproteasome gene missing for PRAAS. J Allergy Clin Immunol. 2020;145(3):1015-1017. [27] XiAO X, FENG X, YOO JH, et al. The β-Grasp Domain of Proteasomal ATPase Mpa Makes Critical Contacts with the Mycobacterium tuberculosis 20S Core Particle to Facilitate Degradation. Msphere. 2022;7(5):e0027422. [28] SHI CX, ZHU YX, BRUINS LA, et al. Proteasome subunits differentially control myeloma cell viability and proteasome inhibitor sensitivity. Mol Cancer Res. 2020; 18(10):1453-1464. [29] GUIMARÃES G, GOMES MT, CAMPOS PC, et al. Immunoproteasome subunits are required for CD8+ T cell function and host resistance to Brucella abortus infection in mice. Infect Immun. 2018;86(3):10-128. [30] WEHR P, PURVIS H, LAW SC, et al. Dendritic cells, T cells and their interaction in rheumatoid arthritis. Clin Exp Immunol. 2019;196(1):12-27. [31] HE J, LI Y, CHEN J, et al. The relationships of CD8+ T cell subsets in RA patients with disease activity and clinical parameters. Int Immunopharmacol. 2023;114:109399. [32] JONSSON AH, ZHANG F, DUNLAP G, et al.Granzyme K+ CD8 T cells form a core population in inflamed human tissue. Sci Transl Med. 2022;14(649):eabo0686. [33] HUANG B, CHEN Z, GENG L, et al. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179(5): 1160-1176. [34] CASALEGNO GARDUÑO R, DÄBRITZ J. New insights on CD8+ T cells in inflammatory bowel disease and therapeutic approaches. Front Immunol. 2021;12:738762. [35] LI CH, MA ZZ, JIAN LL, et al. Iguratimod inhibits osteoclastogenesis by modulating the RANKL and TNF-α signaling pathways. Int Immunopharmacol. 2021;90:107219. [36] CAI L, MU YR, LIU MM, et al. Penta-acetyl Geniposide suppresses migration, invasion, and inflammation of TNF-α-stimulated rheumatoid arthritis fibroblast-like synoviocytes involving wnt/β-catenin signaling pathway. Inflammation. 2021; 44(6):2232-2245. [37] SCHMITT H, NEURATH MF, ATREYA R. Role of the IL23/IL17 Pathway in Crohn’s Disease. Front Immunol. 2021;12:622934. [38] LOH W, VERMEREN S. Anti-inflammatory neutrophil functions in the resolution of inflammation and tissue repair. Cells. 2022;11(24):4076. [39] OLIVEIRA SR, DE ARRUDA JA, SCHNEIDER AH, et al. Neutrophil extracellular traps in rheumatoid arthritis and periodontitis: Contribution of PADI4 gene polymorphisms. J Clin Periodontol. 2024;51(4):452-463. [40] WRIGHT HL, LYON M, CHAPMAN EA, et al.Rheumatoid arthritis synovial fluid neutrophils drive inflammation through production of chemokines, reactive oxygen species, and neutrophil extracellular traps. Front Immunol. 2021;11:584116. [41] GARRIDO-TRIGO A, CORRALIZA AM, VENY M, et al. Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat Commun. 2023;14(1):4506. [42] CHEN H, WU X, XU C, et al. Dichotomous roles of neutrophils in modulating pathogenic and repair processes of inflammatory bowel diseases. Precis Clin Med. 2021;4(4):246-257. [43] WÉRA O, LANCELLOTTI P, OURY C. The dual role of neutrophils in inflammatory bowel diseases. J Clin Med. 2016;5(12):118. [44] SOUFLI I, TOUMI R, RAFA H, et al. Overview of cytokines and nitric oxide involvement in immuno-pathogenesis of inflammatory bowel diseases. World J Gastrointest Pharmacol Ther. 2016;7(3):353. |
| [1] | Lai Jiaming, , Song Yuling, Chen Zixi, Wei Jinghuan, Cai Hao, , Li Guoquan, . Screening of diagnostic markers for endothelial cell Senescence in mice with radiation-induced heart disease and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1450-1463. |
| [2] | Wu Zhilin, , He Qin, Wang Pingxi, Shi Xian, Yuan Song, Zhang Jun, Wang Hao . DYRK2: a novel therapeutic target for rheumatoid arthritis combined with osteoporosis based on East Asian and European populations [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1569-1579. |
| [3] | Lyu Guoqing, Aizimaitijiang·Rouzi, Xiong Daohai. Irisin inhibits ferroptosis in human articular chondrocytes: roles and mechanisms [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(6): 1359-1367. |
| [4] | Zhang Qian, Huang Dongfeng. Weighted gene co-expression network analysis combined with machine learning to screen and validate biomarkers for osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1096-1105. |
| [5] | Liu Kexin, , Hao Kaimin, Zhuang Wenyue, , Li Zhengyi. Autophagy-related gene expression in pulmonary fibrosis models: bioinformatic analysis and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1129-1138. |
| [6] | Zheng Yin, Wu Zhenhua, Zhang Cheng, Ruan Kexin, Gang Xiaolin, Ji Hong. Safety and efficacy of immunoadsorption therapy for rheumatoid arthritis: a network meta-analysis and systematic review [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(5): 1260-1268. |
| [7] | Gu Fucheng, Yang Meixin, Wu Weixin, Cai Weijun, Qin Yangyi, Sun Mingyi, Sun Jian, Geng Qiudong, Li Nan. Effects of Guilu Erxian Glue on gut microbiota in rats with knee osteoarthritis: machine learning and 16S rDNA analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(4): 1058-1072. |
| [8] | Guan Yujie, Zhao Bin. Application and prospect of artificial intelligence in screening and diagnosis of scoliosis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 721-730. |
| [9] | Wang Zhipeng, Zhang Xiaogang, Zhang Hongwei, Zhao Xiyun, Li Yuanzhen, Guo Chenglong, Qin Daping, Ren Zhen. A systematic review of application value of machine learning to prognostic prediction models for patients with lumbar disc herniation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 740-748. |
| [10] | Zhao Feifan, Cao Yujing. An artificial neural network model of ankylosing spondylitis and psoriasis shared genes and machine learning-based mining and validation [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(3): 770-784. |
| [11] | Li Wenhui, Fan Weijing, Liu Guobin. Impact of Zi-Zhu ointment on the miRNA expression profile in mouse models of diabetic ulcers: a high-throughput sequencing analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(17): 4337-4346. |
| [12] | Li Ruiying, Xia Hong. Visual analysis of cuproptosis research: global landscape of hotspots and frontiers [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(17): 4529-4541. |
| [13] | Han Jie, Yao Guojun, Huang Yebao, Xu Zhiwei, Shao Weigang, Shang Kebin, Wu Yachao, Liao Zhen. Genetic structure of co-morbidity between frailty and rheumatoid arthritis: a genome-wide association analysis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(17): 4548-4556. |
| [14] | Chen Feijun, Chen Yingguo, Li Zhengyang, Hu Yuan, Li Fang. Predictive efficacy of machine learning models for postoperative prognosis in older adult patients with acute intracerebral hemorrhage [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(16): 4045-4053. |
| [15] | Wu Jun, Zhang Yuzhu, Dong Xiaojie, Wang Kaidi, Sun Bin. Experimental validation of cytokine-cytokine receptor interaction pathway related gene signatures and molecular subtypes in rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2026, 30(12): 3145-3155. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||