Chinese Journal of Tissue Engineering Research ›› 2021, Vol. 25 ›› Issue (7): 1056-1063.doi: 10.3969/j.issn.2095-4344.2172
Previous Articles Next Articles
Direct reprogramming hepatocytes into islet-like cells by efficiently targeting and activating the endogenous genes
Wang Hanyue1, 2, Li Furong2, Yang Xiaofei1, 2, Hu Chaofeng1
- 1Department of Pathology and Pathophysiology, School of Medicine, Jinan University, Guangzhou 510632, Guangdong Province, China; 2Translational Medicine Collaborative Innovation Center, Second Clinical Medical College (Shenzhen People’s Hospital) of Jinan University, Shenzhen 518020, Guangdong Province, China
-
Received:2020-01-19Revised:2020-01-20Accepted:2020-03-13Online:2021-03-08Published:2020-12-08 -
Contact:Hu Chaofeng, MD, Professor, Department of Pathology and Pathophysiology, School of Medicine, Jinan University, Guangzhou 510632, Guangdong Province, China Yang Xiaofei, MD, Assistant researcher, Department of Pathology and Pathophysiology, School of Medicine, Jinan University, Guangzhou 510632, Guangdong Province, China; Translational Medicine Collaborative Innovation Center, Second Clinical Medical College (Shenzhen People’s Hospital) of Jinan University, Shenzhen 518020, Guangdong Province, China -
About author:Wang Hanyue, Mater candidate, Department of Pathology and Pathophysiology, School of Medicine, Jinan University, Guangzhou 510632, Guangdong Province, China; Translational Medicine Collaborative Innovation Center, Second Clinical Medical College (Shenzhen People’s Hospital) of Jinan University, Shenzhen 518020, Guangdong Province, China -
Supported by:the National Natural Science Foundation for the Youth of China, No. 81800685; the National Natural Science Foundation of China, No. 81670702; the National Natural Science Foundation for the Youth of Guangdong Province, No. 2018A030310039; the Free Exploration Project of Basic Research of Science and Technology Innovation Commission of Shenzhen, No. JCYJ20170307100154602
CLC Number:
Cite this article
Wang Hanyue, Li Furong, Yang Xiaofei, Hu Chaofeng. Direct reprogramming hepatocytes into islet-like cells by efficiently targeting and activating the endogenous genes[J]. Chinese Journal of Tissue Engineering Research, 2021, 25(7): 1056-1063.
share this article
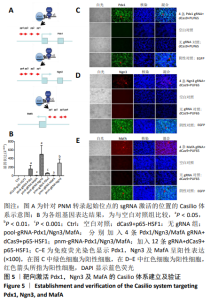
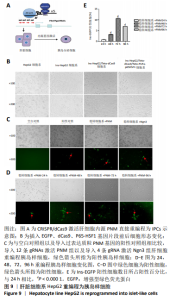
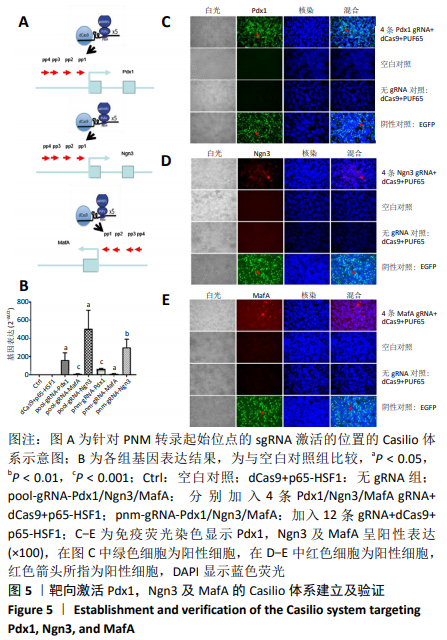
2.1 靶向激活Pdx1,Ngn3及MafA的Casilio体系建立及验证 应用qPCR和免疫荧光检测PNM在293T细胞系中的表达水平,以验证Casilio体系对细胞系内源基因的激活作用。结果显示,与Ctrl组以及No gRNA组相比,分别加入针对Pdx1,Ngn3及MafA的4条gRNA组的基因水平分别上调163倍(P=0.023 2), 503倍(P=0.013 7)和 12倍(P=0.000 2)。同时加入针对PNM的12条gRNA组,Pdx1基因表达水平上调 62倍(P=0.000 2), Ngn3基因表达水平上调301倍(P=0.004 6), MafA基因表达水平上调12倍(P=0.010 8),当加入的gRNA达到12条后,与只加了4 gRNAs组相比基因表达无明显差异(P=0.196 0),见图5A,B。通过免疫荧光检测到了内源PNM蛋白的表达,见图5C-E所示,表明Casilio体系可以实现内源基因的靶向激活。"
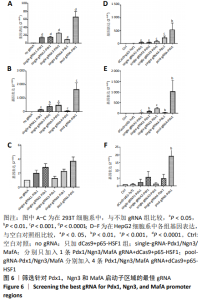
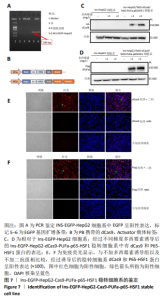
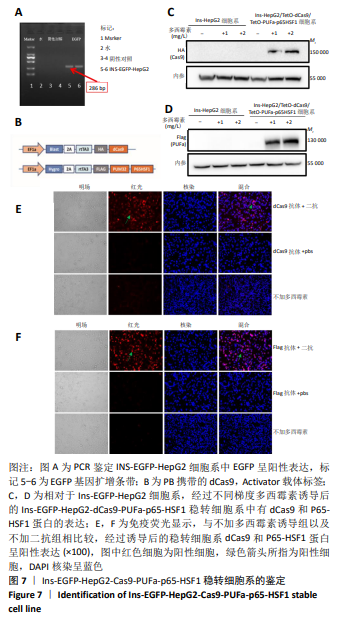
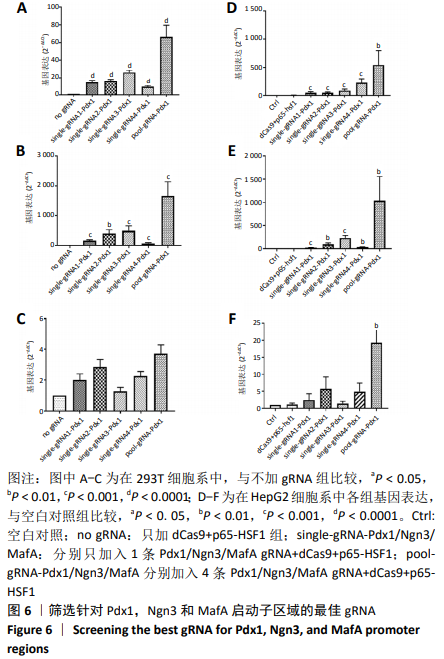
2.2 筛选针对Pdx1,Ngn3,MafA启动子区域的最佳gRNA 为了筛选Casilio体系中激活作用最佳的gRNA,将dCas9、PUFa-P65-HSF1表达质粒与分别针对Pdx1,Ngn3及MafA启动子区域的4条gRNA的表达质粒按照1∶1∶1的比例单独或联合转染293T和HepG2细胞。如图6所示,在293T细胞系中Pdx1基因的gRNA3上调26倍 (P < 0.000 1),gRNA2上调16倍(P < 0.000 1),在4条gRNA中上调倍数最高;针对Ngn3基因的gRNA3和gRNA2上调倍数最高,分别上调501倍(P=0.000 8)和398倍(P=0.001 0);MafA基因gRNA2和gRNA4上调倍数最高,分别上调3倍和2倍。在HepG2细胞系中针对Pdx1基因的gRNA4和 gRNA3上调倍数最高,分别上调223倍(P=0.000 4)和82倍(P=0.000 3);针对Ngn3基因的gRNA3和 gRNA2上调倍数最高,分别上调 234倍(P=0.000 1) 和95倍(P=0.001 1)。针对MafA基因的gRNA2和gRNA4上调倍数最高,分别上调6倍和5倍。针对Pdx1,Ngn3和MafA基因的4条gRNA在293T和HepG2细胞中均有协同效应,即4条gRNA同时转染的激活效率高于单个gRNA转染。 2.3 Ins-EGFP-HepG2-dCas9-PUFa-p65-HSF1稳转细胞系的构建及鉴定 为了验证PNM的重编程效率,利用慢病毒构建针对Insulin基因启动子的绿色荧光报告基因(EGFP)细胞系,用以标记胰岛素基因表达阳性的细胞,见图7A,结果显示EGFP报告基因插入稳转细胞系基因组中。由于dCas9及PUFa-p65-HSF1序列较长,质粒的表达效率低,因此,利用PB转座子体系在Ins-EGFP-HepG2细胞系的基础上,构建了稳定表达dCas9和PUFa-P65-HSF1的Ins-EGFP-HepG2-dCas9-PUFa-p65-HSF1稳转细胞系,PB携带的dCas9和Activator载体标签,见图7B。免疫印迹和免疫荧光结果均显示,经过筛选及多西环素诱导后的稳转细胞系中有dCas9和P65-HSF1蛋白的表达,见图7C-F,表明PB转座子成功实现了dCas9,P65-HSF1片段在HepG2肝细胞系基因组的整合,并且稳转细胞系构建过程中未见细胞形态的明显改变。稳定表达dCas9和PUFa-p65-HSF1蛋白的稳转细胞系只需导入gRNA即可实现内源基因激活作用。"
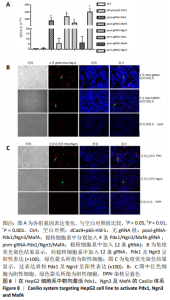
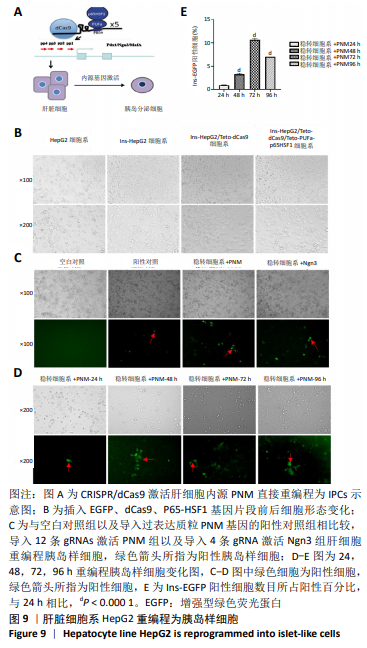
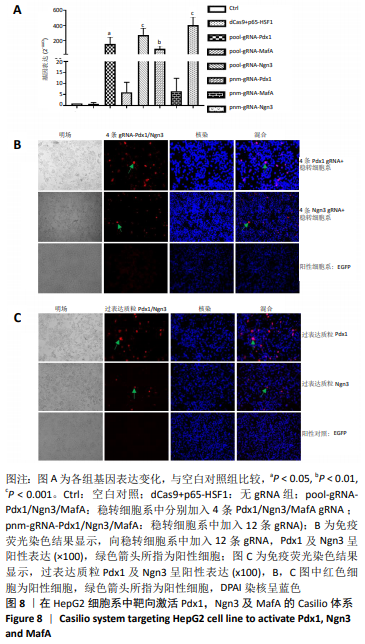
2.4 Casilio体系将肝脏细胞系HepG2重编程为胰岛样细胞 向稳转细胞系中分别导入针对Pdx1,Ngn3及MafA启动子的4条gRNA,qPCR检测内源基因的相对表达水平,结果显示Pdx1基因上调156倍(P=0.014),Ngn3基因上调275倍(P=0.001),MafA基因上调6倍。同时导入针对3个基因的12条gRNA后,Pdx1基因表达上调93倍(P=0.004 6),Ngn3基因表达上调 404倍(P=0.000 3),MafA基因表达上调7倍,见图8A,说明Cailio体系可以高效靶向激活PNM。免疫荧光结果显示,相对于Pdx1及Ngn3过表达质粒,见图8C,Casilio体系可以实现Pdx1及Ngn3蛋白的明显表达,进一步证实了Casilio体系激活内源基因的作用,见图8B。"
| [1] BECK RW, BERGENSTAL RM, LAFFEL LM, et al. Advances in technology for management of type 1 diabetes. Lancet. 2019;394(10205): 1265-1273. [2] BRUNI A, GALA-LOPEZ B, PEPPER AR, et al. Islet cell transplantation for the treatment of type 1 diabetes: recent advances and future challenges. Diabetes Metab Syndr Obes. 2014;7:211. [3] ABREU J, ROEP B. Immune monitoring of islet and pancreas transplant recipients. Curr Diab Rep. 2013;13(5):704-712. [4] KANAK MA, TAKITA M, KUNNATHODI F, et al. Inflammatory response in islet transplantation. Int J Endocrinol. 2014;2014:451035. [5] SHAPIRO AJ, LAKEY JR, RYAN EA, et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N Engl J Med. 2000;343(4):230-238. [6] MANOHAR R, LAGASSE E. Transdetermination: a new trend in cellular reprogramming. Mol Ther. 2009;17(6):936-938. [7] THOWFEEQU S, LI WC, SLACK JM, et al. Reprogramming of liver to pancreas. Methods Mol Biol. 2009;482:407-418. [8] 任丽伟,杨晓菲,李富荣.细胞直接重编程—治疗糖尿病的新技术[J].中国病理生理杂志, 2014;30(12):2284-2288. [9] AKINCI E, BANGA A, TUNGATT K, et al. Reprogramming of various cell types to a beta-like state by Pdx1, Ngn3 and MafA. PLoS One. 2013; 8(11):e82424. [10] BANGA A, AKINCI E, GREDER LV, et al. In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc Natl Acad Sci U S A. 2012;109(38):15336-15341. [11] YECHOOR V, LIU V, ESPIRITU C, et al. Neurogenin3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev Cell. 2009;16(3): 358-373. [12] HICKEY RD, GALIVO F, SCHUG J, et al. Generation of isletlike cells from mouse gall bladder by direct ex vivo reprogramming. Stem Cell Res. 2013;11:503-515. [13] HANNA J, SAHA K, PANDO B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462(7273): 595-601. [14] YOU L, TONG R, LI M, et al. Advancements and obstacles of CRISPR-Cas9 technology in translational research. Mol Ther Methods Clin Dev. 2019;13:359-370. [15] RAPOSO VL. CRISPR-Cas9 and the Promise of a Better Future. Eur J Health Law. 2019;26(4):308-329. [16] GLEESON A, SAWYER A. CRISPR/Cas9: the gold standard of genome editing? Biotechniques. 2018;64(6):239-243. [17] LI D, ZHOU H, ZENG X. Battling CRISPR-Cas9 off-target genome editing. Cell Biol Toxicol. 2019;35(5):403-406. [18] JEHUDA RB, SHEMER Y, BINAH O, et al. Genome editing in induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rev Rep. 2018; 14(3):323-336. [19] CERMAK T, DOYLE EL, CHRISTIAN M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39(12):e82. [20] HOCKEMEYER D, WANG H, KIANI S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8): 731-734. [21] TAGHBALOUT A, DU M, JILLETTE N, et al. Enhanced CRISPR-based DNA demethylation by Casilio-ME-mediated RNA-guided coupling of methylcytosine oxidation and DNA repair pathways. Nat Commun Vol. 2019;10(1):1-12. [22] CHENG AW, WANG H, YANG H, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23(10):1163-1171. [23] SUN S, XIAO J, HUO J, et al. Targeting ectodysplasin promotor by CRISPR/dCas9-effector effectively induces the reprogramming of human bone marrow-derived mesenchymal stem cells into sweat gland-like cells. Stem Cell Res Ther. 2018;9(1):8. [24] HUANG H, ZHONG L, ZHOU J, et al. Leydig-like cells derived from reprogrammed human foreskin fibroblasts by CRISPR/dCas9 increase the level of serum testosterone in castrated male rats. J Cell Mol Med. 2020;24(7):3971-3981. [25] HUANG H, ZOU X, ZHONG L, et al. CRISPR/dCas9-mediated activation of multiple endogenous target genes directly converts human foreskin fibroblasts into Leydig-like cells. J Cell Mol Med. 2019;23(9):6072-6084. [26] LIU P, CHEN M, LIU Y, et al. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 2018;22(2):252-261. [27] ZHOU H, LIU J, ZHOU C, et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR–dCas9-activator transgenic mice. Nat Neurosci. 2018;21(3):440-446. [28] LIAO H-K, HATANAKA F, ARAOKA T, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017; 171(7):1495-1507. [29] KAWAKAMI K, SHIMA A, KAWAKAMI N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97(21):11403-11408. [30] MITRA R, FAIN-THORNTON J, CRAIG NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27(7):1097-1109. [31] CHENG AW, JILLETTE N, LEE P, et al. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016;26(2):254. [32] 王皓毅,李劲松,李伟.基于CRISPR-Cas9新型基因编辑技术研究[J].生命科学, 2016;28(8):867-870. [33] YUSA K, ZHOU L, LI MA, et al. A hyperactive piggyBac transposase for mammalian applications. PNAS. 2011;108(4):1531-1536. [34] PEREZ-PINERA P, KOCAK DD, VOCKLEY CM, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods. 2013;10(10):973. [35] MAEDER ML, LINDER SJ, CASCIO VM, et al. CRISPR RNA–guided activation of endogenous human genes. Nat Methods. 2013;10(10): 977-979. [36] ZHU S, RUSS HA, WANG X, et al. Human pancreatic beta-like cells converted from fibroblasts. Nat Commun. 2016;7(1):1-13. [37] SCHERTZER MD, THULSON E, BRACEROS KC, et al. A piggyBac-based toolkit for inducible genome editing in mammalian cells. RNA. 2019;25(8):1047-1058. [38] HAZELBAKER DZ, BECCARD A, ANGELINI G, et al. A multiplexed gRNA piggyBac transposon system facilitates efficient induction of CRISPRi and CRISPRa in human pluripotent stem cells. Sci Rep. 2020;10(1):1-10. [39] WANG J, HUANG J, XU R. Seamless genome editing in Drosophila by combining CRISPR/Cas9 and piggyBac technologies. Yi Chuan. 2019; 41(5):422-429. |
| [1] | Zhang Yu, Tian Shaoqi, Zeng Guobo, Hu Chuan. Risk factors for myocardial infarction following primary total joint arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1340-1345. |
| [2] | Xiao Guoqing, Liu Xuanze, Yan Yuhao, Zhong Xihong. Influencing factors of knee flexion limitation after total knee arthroplasty with posterior stabilized prostheses [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1362-1367. |
| [3] | Wang Haiying, Lü Bing, Li Hui, Wang Shunyi. Posterior lumbar interbody fusion for degenerative lumbar spondylolisthesis: prediction of functional prognosis of patients based on spinopelvic parameters [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1393-1397. |
| [4] | Yuan Jiawei, Zhang Haitao, Jie Ke, Cao Houran, Zeng Yirong. Underlying targets and mechanism of Taohong Siwu Decoction in prosthetic joint infection on network pharmacology [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1428-1433. |
| [5] | Zhang Chao, Lü Xin. Heterotopic ossification after acetabular fracture fixation: risk factors, prevention and treatment progress [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1434-1439. |
| [6] | Gu Xia, Zhao Min, Wang Pingyi, Li Yimei, Li Wenhua. Relationship between hypoxia inducible factor 1 alpha and hypoxia signaling pathway [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1284-1289. |
| [7] | Wu Xun, Meng Juanhong, Zhang Jianyun, Wang Liang. Concentrated growth factors in the repair of a full-thickness condylar cartilage defect in a rabbit [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1166-1171. |
| [8] | Shen Jinbo, Zhang Lin. Micro-injury of the Achilles tendon caused by acute exhaustive exercise in rats: ultrastructural changes and mechanism [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1190-1195. |
| [9] | Li Jing, Xie Jianshan, Cui Huilin, Cao Ximei, Yang Yanping, Li Hairong. Expression and localization of diacylglycerol kinase zeta and protein kinase C beta II in mouse back skin with different coat colors [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1196-1200. |
| [10] | Tang Hui, Yao Zhihao, Luo Daowen, Peng Shuanglin, Yang Shuanglin, Wang Lang, Xiao Jingang. High fat and high sugar diet combined with streptozotocin to establish a rat model of type 2 diabetic osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1207-1211. |
| [11] | Chai Le, Lü Jianlan, Hu Jintao, Hu Huahui, Xu Qingjun, Yu Jinwei, Quan Renfu. Signal pathway variation after induction of inflammatory response in rats with acute spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1218-1223. |
| [12] | Chen Jiming, Wu Xiaojing, Liu Tianfeng, Chen Haicong, Huang Chengshuo. Effects of silymarin on liver injury and bone metabolism induced by carbon tetrachloride in mice [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1224-1228. |
| [13] | Geng Qiudong, Ge Haiya, Wang Heming, Li Nan. Role and mechanism of Guilu Erxianjiao in treatment of osteoarthritis based on network pharmacology [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1229-1236. |
| [14] | Tan Jingyu, Liu Haiwen. Genome-wide identification, classification and phylogenetic analysis of Fasciclin gene family for osteoblast specific factor 2 [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1243-1248. |
| [15] | Li Zhongfeng, Chen Minghai, Fan Yinuo, Wei Qiushi, He Wei, Chen Zhenqiu. Mechanism of Yougui Yin for steroid-induced femoral head necrosis based on network pharmacology [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(8): 1256-1263. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||