Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (5): 1091-1100.doi: 10.12307/2025.288
Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration
Liu Yani1, Yang Jinghuan2, Lu Huihui1, Yi Yufang1, Li Zhixiang1, Ou Yangfu1, Wu Jingli3, Wei Bing4
- 1Department of General Practice, 2Department of Respiratory and Critical Care Medicine, 4Department of Geriatrics, Affiliated Hospital of Guilin Medical University, Guilin 541001, Guangxi Zhuang Autonomous Region, China; 3College of Computer Science and Engineering, Guangxi Normal University, Guilin 541000, Guangxi Zhuang Autonomous Region, China
-
Received:2024-02-06Accepted:2024-03-15Online:2025-02-18Published:2024-06-04 -
Contact:Liu Yani, Department of General Practice, Affiliated Hospital of Guilin Medical University, Guilin 541001, Guangxi Zhuang Autonomous Region, China -
About author:Liu Yani, Master, Chief physician, Department of General Practice, Affiliated Hospital of Guilin Medical University, Guilin 541001, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China (Regional Science Foundation Project), No. 62366007 (to WJL); Guangxi Natural Science Foundation Project (Surface Project), No. 2022GXNSFAA035625 (to WJL); Guangxi Medical and Health Key Cultivation Discipline Construction Project (to WB); Guangxi Zhuang Autonomous Region Healthcare Commission Self-funded Scientific Research Project, No. Z20190719 (to LYN); Teaching Research and Reform Project of Guilin Medical University (Key Project), No. JG202030 (to LYN); Teaching and Education Research and Reform Project of General Practitioner School of Guilin Medical University (General Project), No. GYQKYB214 (to LYN)
CLC Number:
Cite this article
Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
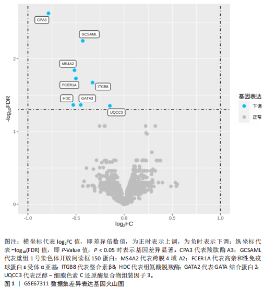
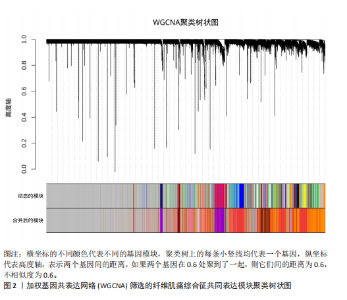
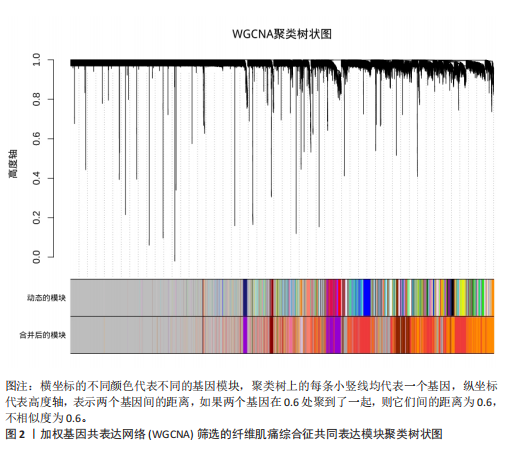
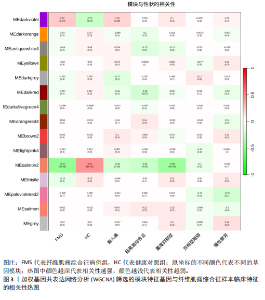
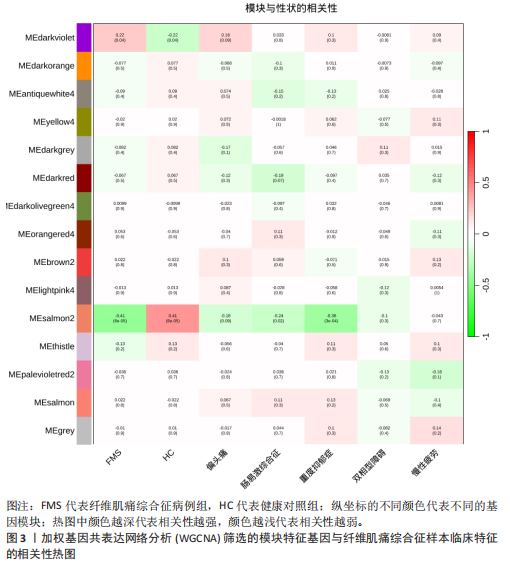
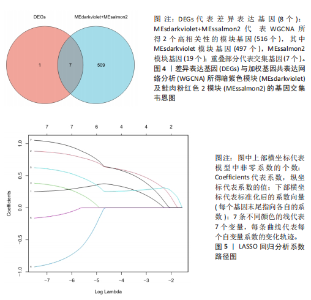
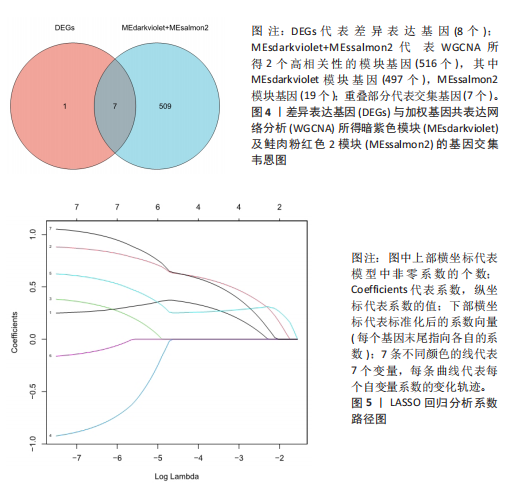
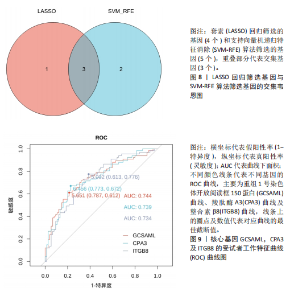
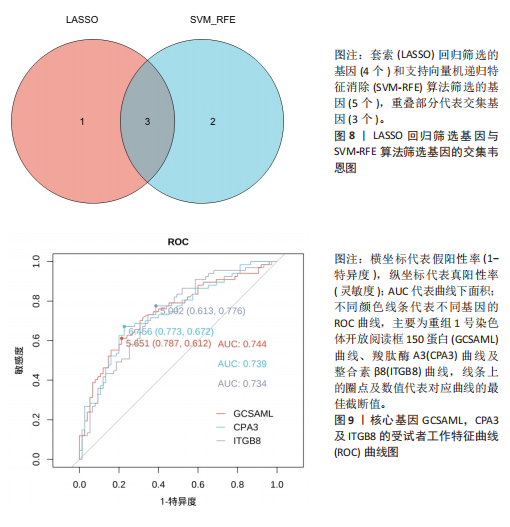
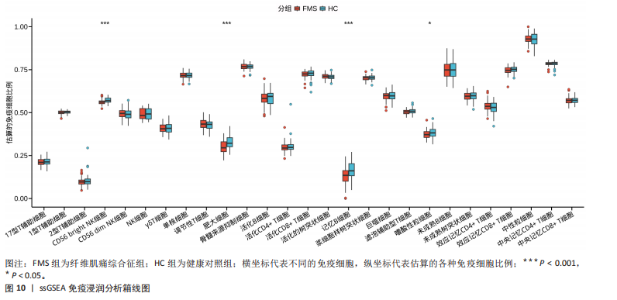
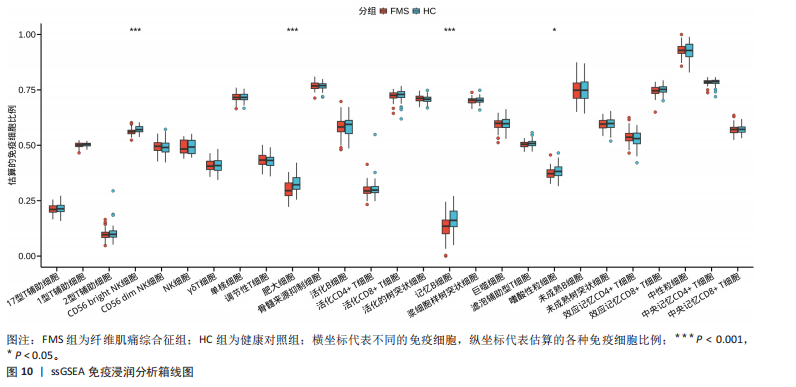
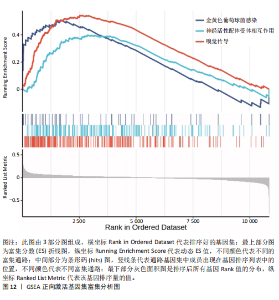
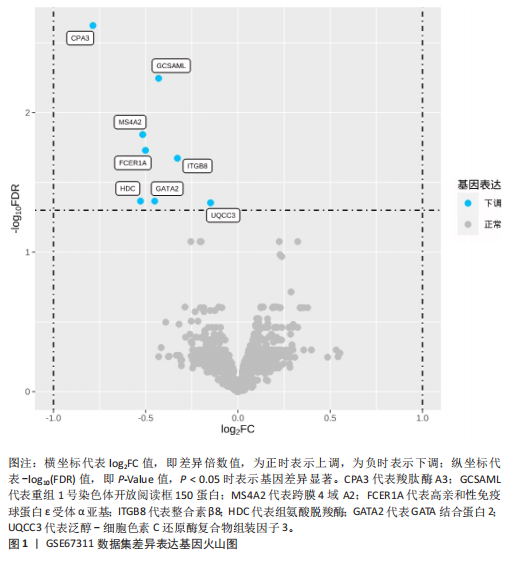
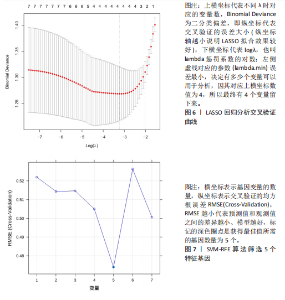
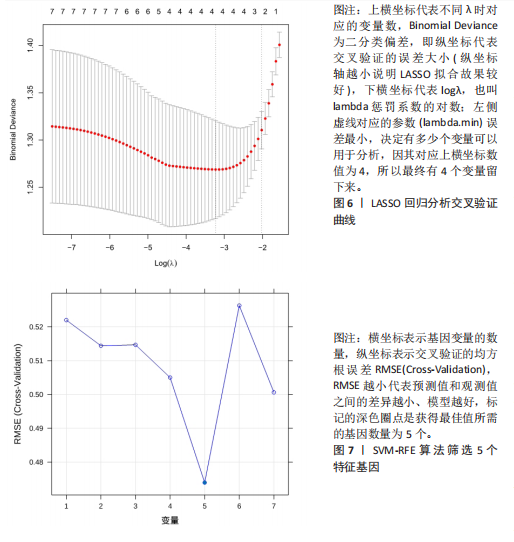
2.1 差异表达基因的筛选结果 通过分析GSE67311数据集中与健康对照组相比纤维肌痛综合征患者外周血标本的转录组测序数据,共筛选出8个差异表达基因,分别为羧肽酶A3(carboxypeptidase A3,CPA3)、重组1号染色体开放阅读框150蛋白 (germinal center associated signaling and motility like,GCSAML)、跨膜4域 A2(membrane-spanning 4-domains subfamily A2,MS4A2)、高亲和性免疫球蛋白ε受体α亚基(Fc fragment of IgE receptor Ia,FCER1A)、整合素β8(Integrin beta-8,ITGB8)、组氨酸脱羧酶(Histidine decarboxylase,HDC)、GATA 结合蛋白2(GATA Binding Protein 2,GATA2)及泛醇-细胞色素C还原酶复合物组装因子3(Ubiquinol-Cytochrome C Reductase Complex Assembly Factor 3,UQCC3),全部为下调基因,绘制的差异火山图详见图1。 2.2 WGCNA及关键模块的识别结果 对数据集GSE67311从1-30的参数值进行WGCNA网络拓扑分析,当R2首次为0.9时软阈值为10,此时邻接函数能够较好满足无尺度条件。接下来,为了保证模块的可靠性,在进行层次聚类时设置剪切高度为35,最小模块基因数目为10,并将剪切高度(MEDissThres)设置为0.6以合并距离相近且相似的模块。最终,经动态分支切割得到15个模块,详见图2。 文章对各模块与临床性状(是否患纤维肌痛综合征)之间的相关性进行进一步分析,其中暗紫色模块(MEdarkviolet)与纤维肌痛综合征的正相关性最高(r=0.22,P=0.04),模块内含有相关基因497个;鲑肉粉红色2模块(MEsalmon2)与纤维肌痛综合征的负相关性最高(r=-0.41,P=6×10-5),模块内含有相关基因19个。另外,对各模块与纤维肌痛综合征临床特征之间的相关性也进行了分析,发现MEsalmon2与纤维肌痛综合征重度抑郁症状之间的负相关性最高(r=-0.38,P=3×10-4),详见图3。 2.3 确定核心基因 差异表达基因与WGCNA分析所得MEsdarkviolet基因及MEsalmon2基因取交集后,获得7个交集基因(FCER1A,CPA3,GCSAML,ITGB8,MS4A2,HDC,GATA2),绘制韦恩图详见图4。进一步进行LASSO回归分析后可获得4个基因(FCER1A,GCSAML,CPA3,ITGB8),详见图5,6;进行SVM-RFE算法分析后可获得5个基因(CPA3,GCSAML,ITGB8,MS4A2,HDC),详见图7。最后,将LASSO回归分析获得的基因与SVM-RFE算法分析获得"
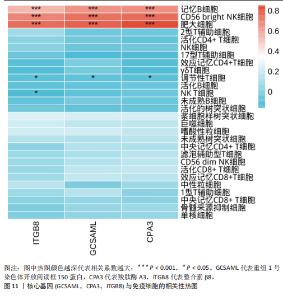
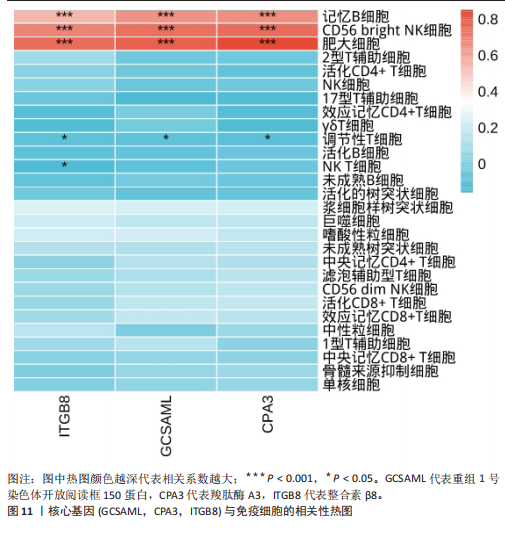
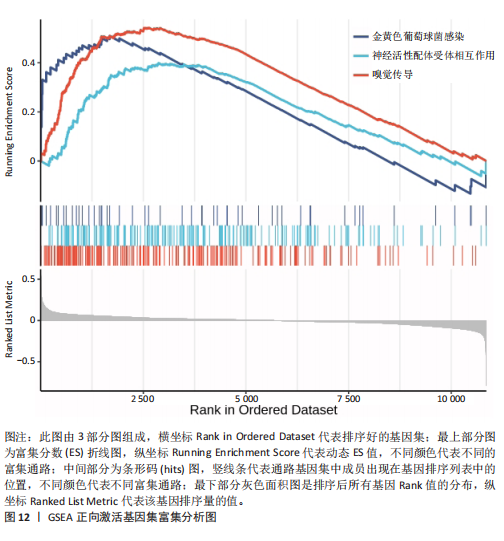
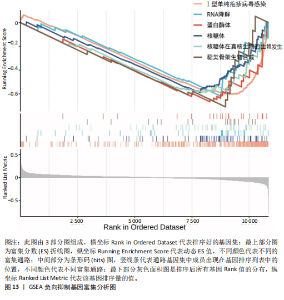
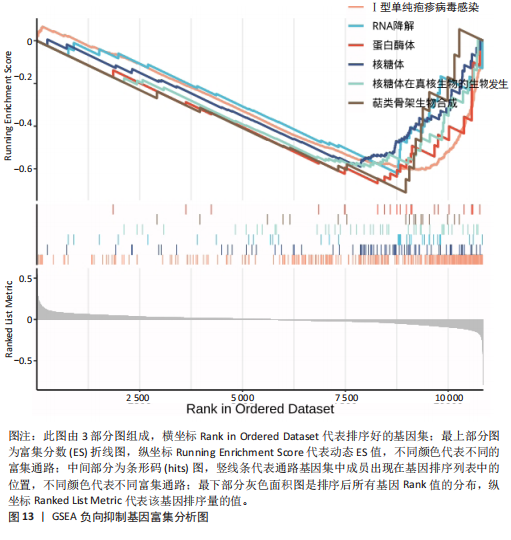
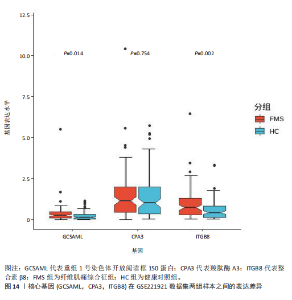
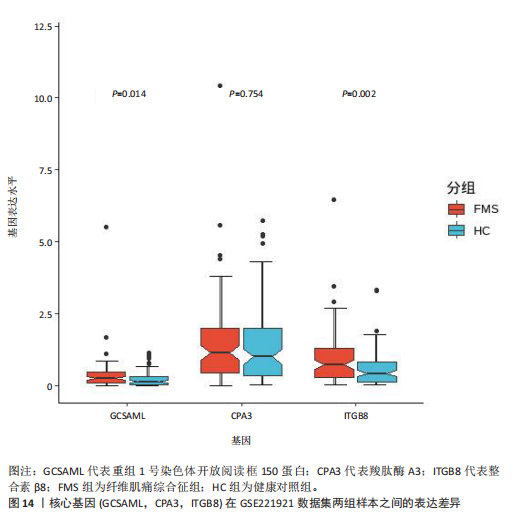
的基因取交集,得到3个核心基因(GCSAML,CPA3,ITGB8),绘制韦恩图详见图8。 2.4 核心基因诊断效能 将筛选出的核心基因GCSAML,CPA3,ITGB8分别绘制ROC曲线,曲线下面积(AUC)分别为0.744,0.739,0.734,均> 0.7,提示可作为生物标记物区分纤维肌痛综合征或健康人,具备良好的诊断效能,详见图9。 2.5 ssGSEA免疫浸润分析结果 ssGSEA免疫浸润分析显示:在纤维肌痛综合征患者外周血中记忆B细胞(Memory B cell)、CD56+ bright NK细胞(CD56 bright natural killer cell)、嗜酸性粒细胞(Eosinophil)、肥大细胞(Mast cell)显著下调,详见图10。免疫细胞与3个核心基因的相关性分析显示:记忆B细胞、CD56 bright NK细胞、肥大细胞与GCSAML,CPA3,ITGB8均呈正相关;调节性T细胞(Regulatory T cell)与GCSAML,CPA3,ITGB8均呈负相关;NK T细胞(Natural killer T cell)与ITGB8呈负相关,详见图11。 2.6 GSEA富集分析结果 GSEA分析结果表明纤维肌痛综合征在9个特征基因集中富集,其中,3条正向激活基因集富集途径,主要与嗅觉传导(Olfactory transduction)、神经活性配体-受体相互作用(Neuroactive ligand-receptor interaction)、金黄色葡萄球菌感染(Staphylococcus aureus infection)信号通路有关,详见图12;6条负向抑制基因集富集途径,分别为1型单纯疱疹病毒感染(Herpes simplex virus 1 infection)、核糖体(Ribosome)、RNA降解(RNA degradation)、核糖体在真核生物中的生物发生 (Ribosome biogenesis in eukaryotes)、蛋白酶体(Proteasome)、萜类骨架生物合成(Terpenoid backbone biosynthesis),详见图13。 2.7 利用外部数据集及文献验证筛选出的核心基因 通过GEO数据库检索,筛选出MOHAPATRA等[8]提交的数据集GSE221921作为外部验证数据集。该研究在芝加哥伊利诺伊大学机构审查委员会批准下进行,以GPL24676为平台,包含96例纤维肌痛综合征患者和93例对照组的外周血样本,均为高通量测序数据。经比较实验所筛选出的3个核心基因(GCSAML,CPA3,ITGB8)在该数据集中纤维肌痛综合征组和对照组中的表达水平,结果发现两组间GCSAML与ITGB8表达水平存在显著差异(P=0.014,P=0.002),CPA3表达水平差异尚未达到显著性意义(P=0.754),绘制的箱线图详见图14。同时,查阅文献验证结果表明,GERRA等[9]在纤维肌痛综合征女性患者的GCSAML基因中发现了6种不同的胞嘧啶甲基化。他们还进一步通过亚硫酸氢盐测序技术对纤维肌痛综合征患者及对照组的11个基因组DNA甲基化水平进行了测序,最终通过比较发现纤维肌痛综合征女性患者体内GCSAML胞嘧啶甲基化水平明显升高,指出GCSAML与纤维肌痛综合征发病存在相关性[10],这与实验发现的GCSAML可作为纤维肌痛综合征核心基因的结论存在一致性。"
| [1] WASTI AZ, MACKAWY AMH, HUSSAIN A, et al. Fibromyalgia interventions, obstacles and prospects: narrative review. Acta Myol. 2023;42(2-3):71-81. [2] FAVRETTI M, IANNUCCELLI C, DI FRANCO M. Pain biomarkers in fibromyalgia syndrome: current understanding and future directions. Int J Mol Sci. 2023;24(13):10443. [3] 刘雅妮,牛红青.25羟基维生素D水平与纤维肌痛综合征关系的Meta分析[J].山西医药杂志,2022,51(2):167-171. [4] LIU W, LI L, YE H, et al. Weighted gene co-expression network analysis in biomedicine research. Shengwu Gongcheng Xuebao. 2017;33(11):1791-1801. [5] LE T, ARONOW RA, KIRSHTEIN A, et al. A review of digital cytometry methods: estimating the relative abundance of cell types in a bulk of cells. Brief Bioinform. 2021;22(4):bbaa219. [6] WANG YY, WANG ZX, HU YD, et al. Current status of pathway analysis in genome-wide association study. Yi Chuan. 2017;39(8): 707-716. [7] JONES KD, GELBART T, WHISENANT TC, et al. Genome-wide expression profiling in the peripheral blood of patients with fibromyalgia. Clin Exp Rheumatol. 2016; 34(2 Suppl 96):S89-S98. [8] MOHAPATRA G, DACHET F, COLEMAN LJ, et al. Identification of unique genomic signatures in patients with fibromyalgia and chronic pain. Sci Rep. 2024;14(1):3949. [9] GERRA MC, CARNEVALI D, PEDERSEN IS, et al. DNA methylation changes in genes involved in inflammation and depression in fibromyalgia: a pilot study. Scand J Pain. 2021;21(2):372-383. [10] GERRA MC, CARNEVALI D, OSSOLA P, et al. DNA methylation changes in fibromyalgia suggest the role of the immune-inflammatory response and central sensitization. J Clin Med. 2021;10(21):4992. [11] FITZCHARLES MA, COHEN SP, CLAUW DJ, et al. Nociplastic pain: towards an understanding of prevalent pain conditions. Lancet. 2021;397(10289):2098-2110. [12] DE TOMMASO M, VECCHIO E, NOLANO M. The puzzle of fibromyalgia between central sensitization syndrome and small fiber neuropathy: a narrative review on neurophysiological and morphological evidence. Neurol Sci. 2022;43(3):1667-1684. [13] JI RR, NACKLEY A, HUH Y, et al. Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology. 2018;129(2):343-366. [14] TSILIONI I, PIPIS H, FREITAG M, et al. Effects of an extract of salmon milt on symptoms and serum TNF and substance p in patients with fibromyalgia syndrome. Clin Ther. 2019;41(8):1564-1574. [15] Al SS, VARGA SJ, Al-HUSINAT L, et al. Unraveling the complex web of fibromyalgia: a narrative review. Medicina (Kaunas). 2024;60(2):272. [16] HELLMAN L, AKULA S, FU Z, et al. Mast cell and basophil granule proteases - in vivo targets and function. Front Immunol. 2022;13:918305. [17] BRUM E, FIALHO M, BECKER G, et al. Involvement of peripheral mast cells in a fibromyalgia model in mice. Eur J Pharmacol. 2024;967:176385. [18] MOKHEMER SA, DESOUKY MK, ABDELGHANY AK, et al. Stem cells therapeutic effect in a reserpine-induced fibromyalgia rat model: a possible NLRP3 inflammasome modulation with neurogenesis promotion in the cerebral cortex. Life Sci. 2023;325:121784. [19] KRUPA AJ, CHROBAK AA, SOLTYS Z, et al. Psychopathological symptoms in fibromyalgia and their associations with resistance to pharmacotherapy with SNRI. Psychiatr Pol. 2024. doi: 10.12740/PP/OnlineFirst/176000. [20] ZABIHIYEGANEH M, AMINI KA, AKBARI A, et al. Association of serum vitamin D status with serum pro-inflammatory cytokine levels and clinical severity of fibromyalgia patients. Clin Nutr ESPEN. 2023;55:71-75. [21] ELLERGEZEN P, ALP A, ÇAVUN S, et al. Pregabalin inhibits proinflammatory cytokine release in patients with fibromyalgia syndrome. Arch Rheumatol. 2023;38(2):307-314. [22] HALILI A. Temporal model for central sensitization: a hypothesis for mechanism and treatment using systemic manual therapy, a focused review. MethodsX. 2023; 10:101942. [23] BANFI G, DIANI M, PIGATTO PD, et al. T Cell Subpopulations in the physiopathology of fibromyalgia: evidence and perspectives. Int J Mol Sci. 2020;21(4):1186. [24] CONTI P, GALLENGA CE, CARAFFA A, et al. Impact of mast cells in fibromyalgia and low-grade chronic inflammation: can IL-37 play a role? Dermatol Ther. 2020;33(1):e13191. [25] THEOHARIDES TC, TSILIONI I, BAWAZEER M. Mast cells, neuroinflammation and pain in fibromyalgia syndrome. Front Cell Neurosci. 2019;13:353. [26] LENERT ME, SZABO-PARDI TA, BURTON MD. Regulatory T-cells and IL-5 mediate pain outcomes in a preclinical model of chronic muscle pain. Mol Pain. 2023;19:804311971. [27] EGE F, ISIK R. A comparative assessment of the inflammatory markers in patients with fibromyalgia under duloxetine treatment. Front Biosci (Landmark Ed). 2023;28(8):161. [28] VALENCIA C, FATIMA H, NWANKWO I, et al. A correlation between the pathogenic processes of fibromyalgia and irritable bowel syndrome in the middle-aged population: a systematic review. Cureus. 2022;14(10):e29923. [29] VERMA V, DRURY GL, PARISIEN M, et al. Unbiased immune profiling reveals a natural killer cell-peripheral nerve axis in fibromyalgia. Pain. 2022;163(7):e821-e836. [30] BLANCO S, SANROMAN L, PEREZ-CALVO S, et al. Olfactory and cognitive functioning in patients with fibromyalgia. Psychol Health Med. 2019;24(5):530-541. [31] SAYILIRS S, ÇULLU N. Decreased olfactory bulb volumes in patients with fibromyalgia syndrome. Clin Rheumatol. 2017;36(12): 2821-2824. [32] ÖZSOY-ÜNÜBOL T, KULLAKÇI H, ILHAN İ, et al. Evaluation of olfactory and gustatory functions in patients with fibromyalgia syndrome: its relationship with anxiety, depression, and alexithymia. Arch Rheumatol. 2020;35(4):584-591. [33] DORRIS ER, MACCARTHY J, SIMPSON K, et al. Sensory perception quotient reveals visual, scent and touch sensory hypersensitivity in people with fibromyalgia syndrome. Front Pain Res (Lausanne). 2022;3:926331. [34] CLAUW D, SARZI-PUTTINI P, PELLEGRINO G, et al. Is fibromyalgia an autoimmune disorder? Autoimmun Rev. 2023. doi: 10.1016/j.autrev.2023.103424. |
| [1] | Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. The construction and validation of a prediction model based on multiple machine learning algorithms and the immunomodulatory analysis of rheumatoid arthritis from the perspective of mitophagy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-15. |
| [2] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [3] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [4] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [5] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [6] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [7] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [8] | Yang Bin, Tao Guangyi, Yang Shun, Xu Junjie, Huang Junqing . Visualization analysis of research hotspots of artificial intelligence in field of spinal cord nerve injury and repair [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(4): 761-770. |
| [9] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| [10] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [11] | Jiang Qiyu, Zeng Huiyan. A novel analysis and prediction method for potential mechanisms of traditional Chinese medicine based on artificial intelligence and omics data-driven approach [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7552-7561. |
| [12] | Chen Ying, Guo Xiaojing, Mo Xueni, Ma Wei, Wu Shangzhi, Li Xiangling, Xie Tingting. Analysis of diagnostic biomarkers for ischemic stroke and experimental validation of targeted cuproptosis related genes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7562-7570. |
| [13] | Gong Yuehong, Wang Mengjun, Ren Hang, Zheng Hui, Sun Jiajia, Liu Junpeng, Zhang Fei, Yang Jianhua, Hu Junping. Machine learning combined with bioinformatics screening of key genes for pulmonary fibrosis associated with cellular autophagy and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7679-7689. |
| [14] | Han Jie, Pan Chengzhen, Shang Yuzhi, Zhang Chi. Identification of immunodiagnostic biomarkers and drug screening for steroid-induced osteonecrosis of the femoral head [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7690-7700. |
| [15] | Li Jing, Lu Guangqi, Zhuang Minghui, Cui Ying, Yu Zhangjingze, Sun Xinyue, Ma Mingming, Zhu Liguo, Yu Jie. Development of a clinical prediction model for cervical instability in young and middle-aged adults based on machine learning [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(33): 7203-7210. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||