Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (32): 6893-6904.doi: 10.12307/2025.912
Previous Articles Next Articles
Oxidative stress-related genes and molecular mechanisms after spinal cord injury: data analysis and verification based on GEO database
Wang Ziheng, Wu Shuang
- Department of Rehabilitation Medicine, Affiliated Hospital of Guizhou Medical University, Guiyang 550000, Guizhou Province, China
-
Received:2024-07-13Accepted:2024-10-31Online:2025-11-18Published:2025-04-26 -
Contact:Wu Shuang, Doctoral supervisor, Chief physician, Department of Rehabilitation Medicine, Affiliated Hospital of Guizhou Medical University, Guiyang 550000, Guizhou Province, China -
About author:Wang Ziheng, MS, Department of Rehabilitation Medicine, Affiliated Hospital of Guizhou Medical University, Guiyang 550000, Guizhou Province, China -
Supported by:National Natural Science Foundation of China (Regional Project), Nos. 82060419 and 82260452 (both to WS); Guizhou Provincial Science and Technology Plan Project, No. ZK(2022)045 (to WS)
CLC Number:
Cite this article
Wang Ziheng, Wu Shuang. Oxidative stress-related genes and molecular mechanisms after spinal cord injury: data analysis and verification based on GEO database[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6893-6904.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

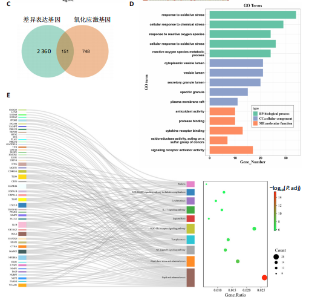
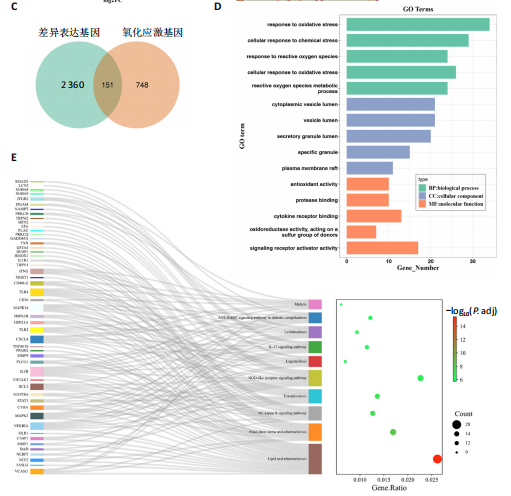
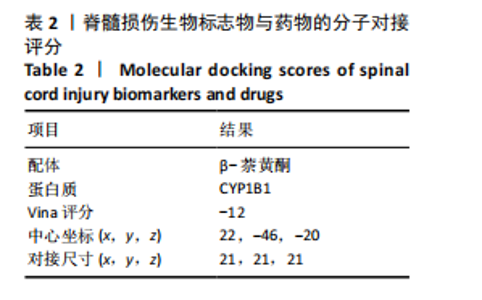
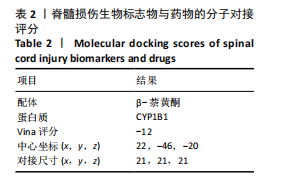
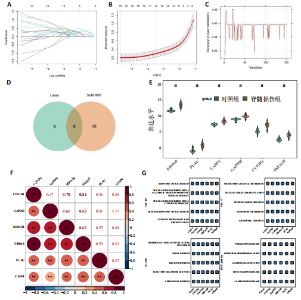
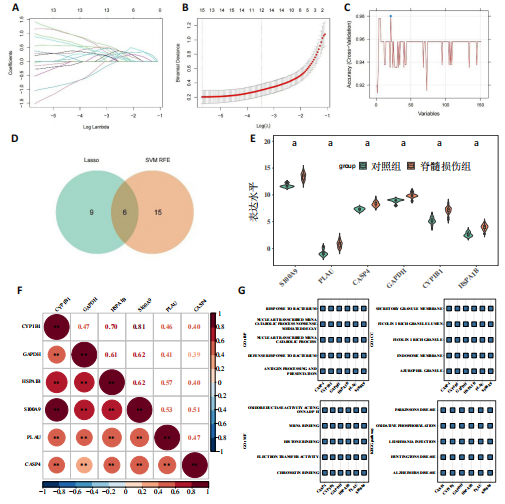
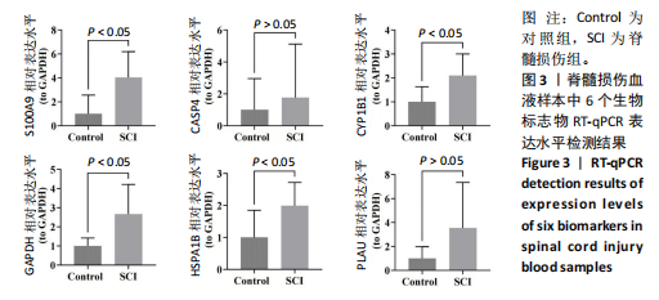
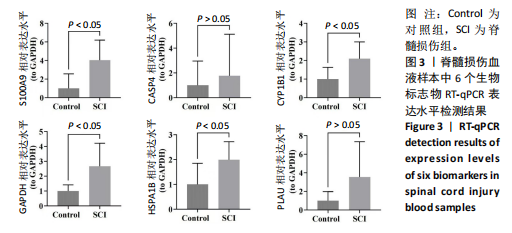
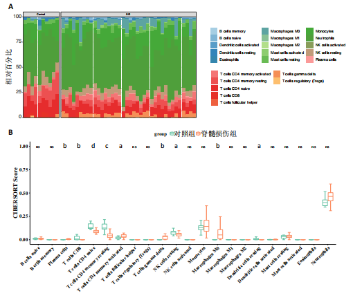
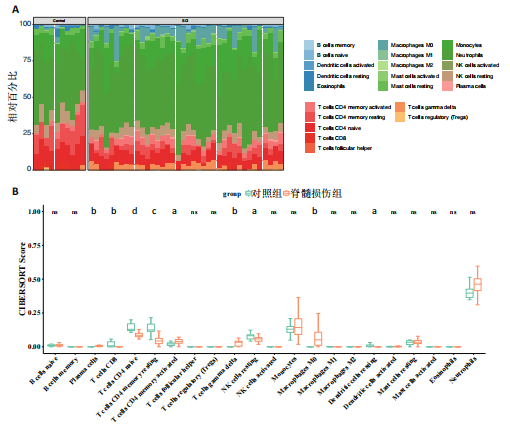
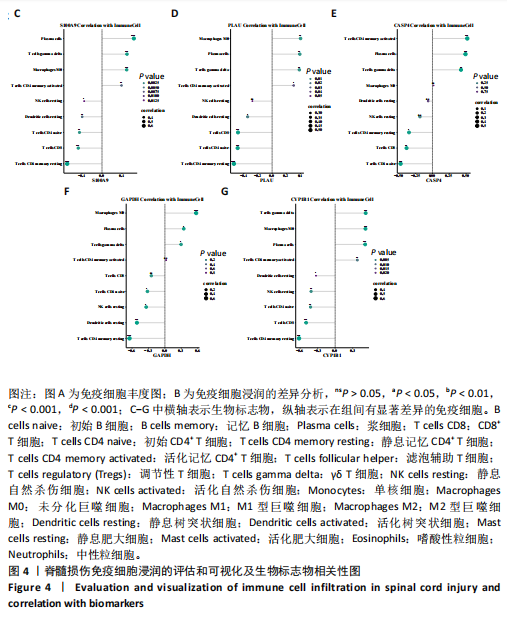
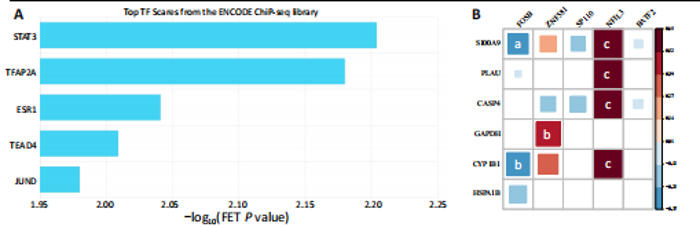
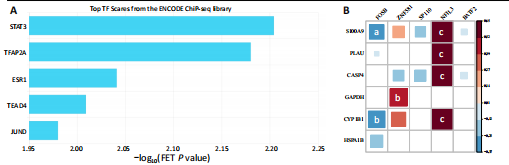
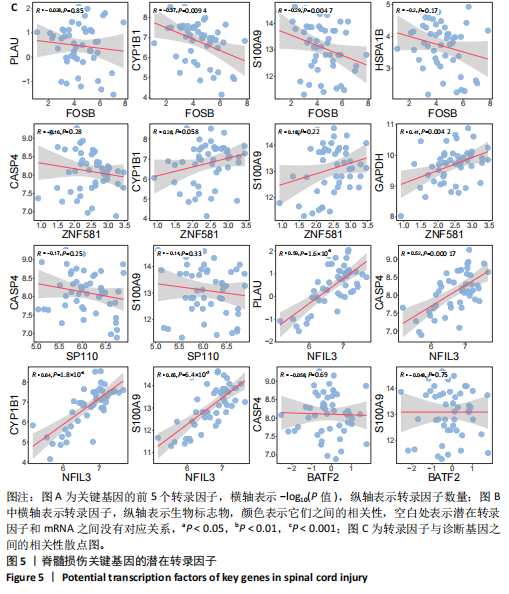
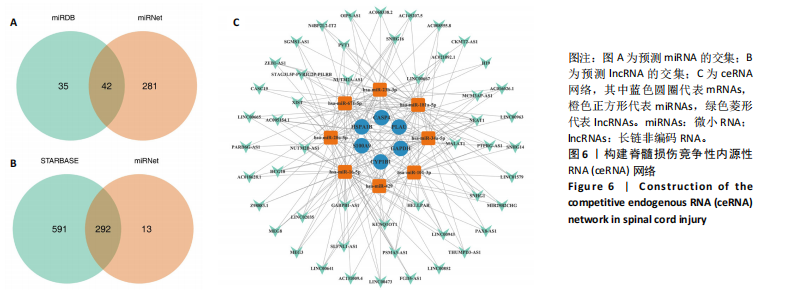
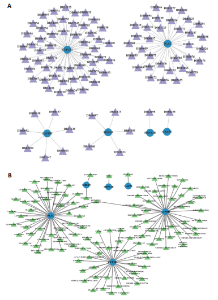
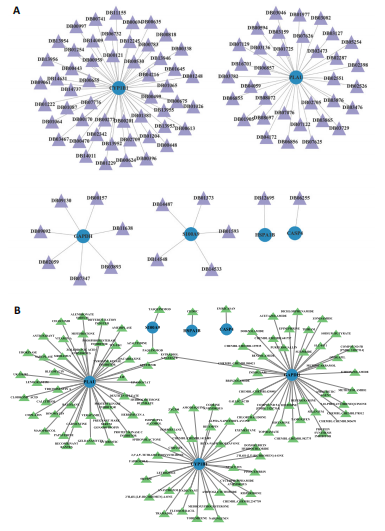
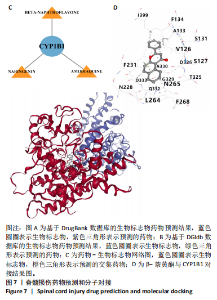
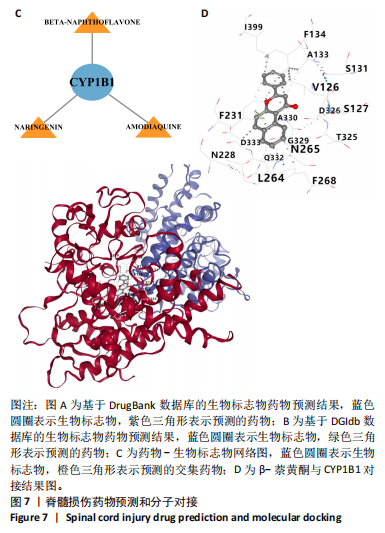
2.1 与脊髓损伤紧密关联的氧化应激相关基因的鉴定和功能注释分析 从GSE151371数据库(脊髓损伤和对照样本)中筛选出2 511个差异表达基因,其中包括1 236个上调基因和1 275个下调基因(图1A,B)。将氧化应激基因与差异表达基因相交,得到了151个与脊髓损伤紧密关联的氧化应激相关基因(图1C)。然后,对151个与脊髓损伤紧密关联的氧化应激相关基因进行GO和KEGG富集分析,结果显示与脊髓损伤紧密关联的氧化应激相关基因参与了1 606个GO条目,包括抗氧化活性、过氧化物质基质、对氧化应激的响应、对活性氧的响应、活性氧代谢过程等(图1D)。与脊髓损伤紧密关联的氧化应激相关基因富集在104个KEGG通路中,包括化学致癌-活性氧、神经营养因子信号通路、多种疾病神经退行性通路等(图1E)。 2.2 生物标志物的获得和分析 使用glment包对151个基因进行LASSO和SVM-RFE分析,分别获得了15和21个基因(图2A-C),将LASSO和SVM-RFE获得的基因取交集获得了6个生物标志物,分别是S100A9、PLAU、CASP4、GAPDH、CYP1B1和HSPA1B(图2D)。从箱线图中可以看出,所有生物标志物在脊髓损伤组和对照组之间的表达差异显著(图2E)。同样,RT-qPCR结果显示,脊髓损伤组和对照组之间S100A9、GAPDH、CYP1B1和HSPA1B存在显著差异,而PLAU和CASP4在脊髓损伤中呈上升趋势(图3)。相关性分析表明,生物标志物之间呈显著正相关,其中S100A9和CYP1B1的正相关性最高(图2F)。单基因集富集分析显示,生物标志物共同参与的GO条目包括抗原处理与呈递、内体膜、作用于烟酰胺腺嘌呤二核苷酸磷酸(nicotinamide adenine dinucleotide phosphate,NADPH)的氧化还原酶活性、mRNA结合、组蛋白结合、电子转移活性等,KEGG通路包括氧化磷酸化等(图2G)。 2.3 免疫浸润分析 为了展示训练集中脊髓损伤和对照组之间免疫细胞含量的差异,构建了免疫细胞浸润水平丰度,如图4A所示,22种免疫细胞在脊髓损伤和对照样本之间存在差异。在两组之间,9种免疫细胞的浸润水平显著不同,包括浆细胞、休息状态的自然杀伤细胞和M0型巨噬细胞等(图4B)。之后分析了基因与差异细胞的相关性,结果表明,6个生物标志物与浆细胞和γδT细胞呈显著正相关,与记忆休息状态的CD4 T细胞和静止状态的CD4 T细胞呈负相关(图4C)。 2.4 转录因子生物标志物的鉴定 为了研究生物标志物的上游调控机制,使用spearman分析构建基因与转录因子的相关性。预测的前5个转录因子为FOSB、ZNF581、SP110、NFIL3和BATF2,其中FOSB、ZNF581和NFIL3与部分生物标志物显著相关,例如,FOSB与S100A9、CYP1B1呈显著负相关,而NFIL3与S100A9、PLAU、CASP4和CYP1B1呈显著正相关(图5A-C)。 2.5 mRNA-miRNA-lncRNA网络及药物预测 为了揭示不同RNA分子之间通过竞争性结合miRNA来调控彼此表达的复杂网络,此次研究从miRDB和miRNet数据库预测的miRNAs中取交集后得到了42个相交的miRNAs(其中有8个miRNAs的调控目标数量大于2),见图6A。通过对StarBase和LncBase Predicted数据库预测的lncRNAs取交集,获得了292个相交的lncRNAs,其中包括50个调控目标数量大于2的lncRNAs(图6B)。然后用6个mRNAs、8个miRNAs和50个lncRNAs形成了mRNA-miRNA-lncRNA网络,包括S100A9-hsa-mir-16-5p-HCG18、CYP1B1-hsa-mir-429-KCNQ1OT1、CASP4-hsa-mir-23b-3p-SNHG16等关系对(图6C)。此外,基于生物标志物,在DrugBank 数据库预测了100种小分子药物,并且它们形成的相互作用对包括DB01373-S100A9、DB01065-CYP1B1和DB00594-PLAU等(图7A)。在DGIdb数据库预测了119个小分析药物(图7B),将2个数据库6个生物标志物预测的药物分别取交集,只有CYP1B1取到了3个交集药物,分别是β-萘黄酮(BETA-NAPHTHOFLAVONE)、阿莫地喹(AMODIAQUINE)、"
| [1] STERNER RC, STERNER RM. Immune response following traumatic spinal cord injury: Pathophysiology and therapies. Front Immunol. 2022;13:1084101. [2] AHUJA CS, WILSON JR, NORI S, et al. Traumatic spinal cord injury. Nat Rev Dis Primers. 2017;3:17018. [3] GUHA L, KUMAR H. Drug Repurposing for Spinal Cord Injury: Progress Towards Therapeutic Intervention for Primary Factors and Secondary Complications. Pharmaceut Med. 2023;37(6):463-490. [4] ROPPER AE, ROPPER AH. Acute Spinal Cord Compression. N Engl J Med. 2017;376(14):1358-1369. [5] GUHA L, KUMAR H. Drug Repurposing for Spinal Cord Injury: Progress Towards Therapeutic Intervention for Primary Factors and Secondary Complications. Pharmaceut Med. 2023;37(6):463-490. [6] MA H, WANG C, HAN L, et al. Tofacitinib Promotes Functional Recovery after Spinal Cord Injury by Regulating Microglial Polarization via JAK/STAT Signaling Pathway. Int J Biol Sci. 2023;19(15):4865-4882. [7] FREYERMUTH-TRUJILLO X, SEGURA-URIBE JJ, SALGADO-CEBALLOS H, et al. Inflammation: A Target for Treatment in Spinal Cord Injury. Cells. 2022;11(17):2692. [8] LI SS, ZHANG BY, YIN SG, et al. A new peptide, VD11, promotes structural and functional recovery after spinal cord injury. Neural Regen Res. 2023;18(10):2260-2267. [9] HAMANN K, SHI R. Acrolein scavenging: a potential novel mechanism of attenuating oxidative stress following spinal cord injury. J Neurochem. 2009;111(6):1348-1356. [10] BRAUGHLER JM, HALL ED. Central nervous system trauma and stroke. I. Biochemical considerations for oxygen radical formation and lipid peroxidation. Free Radic Biol Med. 1989;6(3):289-301. [11] PAPASTEFANAKI F, MATSAS R. From demyelination to remyelination: the road toward therapies for spinal cord injury. Glia. 2015;63(7): 1101-1125. [12] YU Q, JIANG X, LIU X, et al. Glutathione-modified macrophage-derived cell membranes encapsulated metformin nanogels for the treatment of spinal cord injury. Biomater Adv. 2022;133:112668. [13] ZHANG C, ZHAI T, ZHU J, et al. Research Progress of Antioxidants in Oxidative Stress Therapy after Spinal Cord Injury. Neurochem Res. 2023;48(12):3473-3484. [14] FAKHRI S, ABBASZADEH F, MORADI SZ, et al. Effects of Polyphenols on Oxidative Stress, Inflammation, and Interconnected Pathways during Spinal Cord Injury. Oxid Med Cell Longev. 2022;2022: 8100195. [15] WANG B, HUANG M, SHANG D, et al. Mitochondrial Behavior in Axon Degeneration and Regeneration. Front Aging Neurosci. 2021;13: 650038. [16] LI Q, GAO S, KANG Z, et al. Rapamycin Enhances Mitophagy and Attenuates Apoptosis After Spinal Ischemia-Reperfusion Injury. Front Neurosci. 2018;12:865. [17] GU C, LI L, HUANG Y, et al. Salidroside Ameliorates Mitochondria-Dependent Neuronal Apoptosis after Spinal Cord Ischemia-Reperfusion Injury Partially through Inhibiting Oxidative Stress and Promoting Mitophagy. Oxid Med Cell Longev. 2020;2020:3549704. [18] KYRITSIS N, TORRES-ESPIN A, SCHUPP PG, et al. Diagnostic blood RNA profiles for human acute spinal cord injury. J Exp Med. 2021; 218(3):e20201795. [19] LIU S, WANG Z, ZHU R, et al. Three Differential Expression Analysis Methods for RNA Sequencing: limma, EdgeR, DESeq2. J Vis Exp. 2021; (175). doi: 10.3791/62528. [20] WANG F, LIN H, SU Q, et al. Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma. World J Surg Oncol. 2022;20(1):275. [21] XIE L, HUANG G, GAO M, et al. Identification of Atrial Fibrillation-Related lncRNA Based on Bioinformatic Analysis. Dis Markers. 2022; 2022:8307975. [22] YU G, WANG LG, HAN Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5): 284-287. [23] ANJUM A, YAZID MD, FAUZI DM, et al. Spinal Cord Injury: Pathophysiology, Multimolecular Interactions, and Underlying Recovery Mechanisms. Int J Mol Sci. 2020;21(20). doi: 10.3390/ijms21207533. [24] QUADRI S A, FAROOQUI M, IKRAM A, et al. Recent update on basic mechanisms of spinal cord injury. Neurosurg Rev. 2020;43(2):425-441. [25] LIU G, DENG B, HUO L, et al. Temporal profiling and validation of oxidative stress-related genes in spinal cord injury. Brain Res Bull. 2023;205:110832. [26] NAGAREDDY PR, MURPHY AJ, STIRZAKER RA, et al. Hyperglycemia promotes myelopoiesis and impairs the resolution of atherosclerosis. Cell Metab. 2013;17(5):695-708. [27] COTOI OS, DUNER P, KO N, et al. Plasma S100A8/A9 correlates with blood neutrophil counts, traditional risk factors, and cardiovascular disease in middle-aged healthy individuals. Arterioscler Thromb Vasc Biol. 2014;34(1):202-210. [28] PEI H, QU J, CHEN J, et al. S100A9 as a Key Myocardial Injury Factor Interacting with ATP5 Exacerbates Mitochondrial Dysfunction and Oxidative Stress in Sepsis-Induced Cardiomyopathy. J Inflamm Res. 2024;17:4483-4503. [29] FAN S, ZHAO H, LIU Y, et al. Isoproterenol Triggers ROS/P53/S100-A9 Positive Feedback to Aggravate Myocardial Damage Associated with Complement Activation. Chem Res Toxicol. 2020;33(10):2675-2685. [30] MCMAHON BJ, KWAAN HC. Components of the Plasminogen-Plasmin System as Biologic Markers for Cancer. Adv Exp Med Biol. 2015;867: 145-156. [31] MAHMOOD N, MIHALCIOIU C, RABBANI SA. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front Oncol. 2018;8:24. [32] STAN SD, HAHM ER, WARIN R, et al. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 2008;68(18):7661-7669. [33] QU B, JIA Y, LIU Y, et al. The detection and role of heat shock protein 70 in various nondisease conditions and disease conditions: a literature review. Cell Stress Chaperones. 2015;20(6): 885-892. [34] ZININGA T, RAMATSUI L, SHONHAI A. Heat Shock Proteins as Immunomodulants. Molecules. 2018;23(11). doi: 10.3390/molecules 23112846. [35] PISHESHA N, HARMAND TJ, PLOEGH HL. A guide to antigen processing and presentation. Nat Rev Immunol. 2022;22(12):751-764. [36] CHAKRABORTY P, BJORK P, KALLBERG E, et al. Vesicular Location and Transport of S100A8 and S100A9 Proteins in Monocytoid Cells. PLoS One. 2015;10(12):e0145217. [37] CATALA A, ZVARA A, PUSKAS LG, et al. Melatonin-induced gene expression changes and its preventive effects on adriamycin-induced lipid peroxidation in rat liver. J Pineal Res. 2007;42(1):43-49. [38] LIDIN E, SKOLD MK, ANGERIA M, et al. Hippocampal Expression of Cytochrome P450 1B1 in Penetrating Traumatic Brain Injury. Int J Mol Sci. 2022;23(2):722. [39] SHIMADA T, NAGAYOSHI H, MURAYAMA N, et al. Oxidation of 3’-methoxyflavone, 4’-methoxyflavone, and 3’,4’-dimethoxyflavone and their derivatives having 5,7-dihydroxyl moieties by human cytochromes P450 1B1 and 2A13. Xenobiotica. 2022;52(2):134-145. [40] Du F, DING Z, RONNOW CF, et al. S100A9 induces reactive oxygen species-dependent formation of neutrophil extracellular traps in abdominal sepsis. Exp Cell Res. 2022;421(2):113405. [41] MIHAILA AC, CIORTAN L, MACARIE RD, et al. Transcriptional Profiling and Functional Analysis of N1/N2 Neutrophils Reveal an Immunomodulatory Effect of S100A9-Blockade on the Pro-Inflammatory N1 Subpopulation. Front Immunol. 2021;12:708770. [42] HUANG S, ZHEN Y, YIN X, et al. KMT2C Induced by FABP5P3 Aggravates Keratinocyte Hyperproliferation and Psoriasiform Skin Inflammation by Upregulating the Transcription of PIK3R3. J Invest Dermatol. 2023; 143(1):37-47. [43] LIU Q, FU H, SUN F, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36(16): 5391-5404. [44] GONZALEZ-LOPEZ P, ALVAREZ-VILLARREAL M, RUIZ-SIMON R, et al. Role of miR-15a-5p and miR-199a-3p in the inflammatory pathway regulated by NF-kappaB in experimental and human atherosclerosis. Clin Transl Med. 2023;13(8):e1363.
[45] D’ANTONA S, PORRO D, GALLIVANONE F, et al. Characterization of cell cycle, inflammation, and oxidative stress signaling role in non-communicable diseases: Insights into genetic variants, microRNAs and pathways. Comput Biol Med. 2024;174:108346. [46] ZHANG Y, YANG G, LUO Y. Long non-coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR-200a. Exp Ther Med. 2019;17(2):1337-1345. [47] ZHAO W, ZHANG Y, ZHANG M, et al. Effects of total glucosides of paeony on acute renal injury following ischemia-reperfusion via the lncRNA HCG18/miR-16-5p/Bcl-2 axis. Immunobiology. 2022;227(2): 152179. [48] ZOTTA A, O’NEILL L, YIN M. Unlocking potential: the role of the electron transport chain in immunometabolism. Trends Immunol. 2024;45(4): 259-273. [49] HANNAN MA, DASH R, SOHAG A, et al. Neuroprotection Against Oxidative Stress: Phytochemicals Targeting TrkB Signaling and the Nrf2-ARE Antioxidant System. Front Mol Neurosci. 2020;13:116. [50] ZHENG M, LIU Y, ZHANG G, et al. The Applications and Mechanisms of Superoxide Dismutase in Medicine, Food, and Cosmetics. Antioxidants (Basel). 2023;12(9):1675. [51] ANKENY DP, POPOVICH PG. Mechanisms and implications of adaptive immune responses after traumatic spinal cord injury. Neuroscience. 2009;158(3):1112-1121. [52] GARCIA E, AGUILAR-CEVALLOS J, SILVA-GARCIA R, et al. Cytokine and Growth Factor Activation In Vivo and In Vitro after Spinal Cord Injury. Mediators Inflamm. 2016;2016:9476020. [53] XU Y, LI Y, ZHANG Z, et al. [Expressions and implications of Th1/Th2 cytokines in injured rat brain and spinal cord]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2013;29(9):919-922,926. [54] POPOVICH PG, TOVAR CA, WEI P, et al. A reassessment of a classic neuroprotective combination therapy for spinal cord injured rats: LPS/pregnenolone/indomethacin. Exp Neurol. 2012;233(2):677-685. [55] PENG H, TIAN Z. Natural Killer Cell Memory: Progress and Implications. Front Immunol. 2017;8:1143. [56] JUELKE K, KILLIG M, LUETKE-EVERSLOH M, et al. CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood. 2010;116(8):1299-1307. [57] XU L, ZHANG Y, ZHANG R, et al. Elevated plasma BDNF levels are correlated with NK cell activation in patients with traumatic spinal cord injury. Int Immunopharmacol. 2019;74:105722. [58] ZELTER M, MENSCH-DECHENE J, LOCKHART A. [Pulmonary blood volume. Definition, measurement, normal values]. Pathol Biol (Paris). 1975;23(4):323-332. [59] FELTEN DL, FELTEN SY, CARLSON SL, et al. Noradrenergic and peptidergic innervation of lymphoid tissue. J Immunol. 1985; 135(2 Suppl):755s-765s. [60] HERMAN P, STEIN A, GIBBS K, et al. Persons with Chronic Spinal Cord Injury Have Decreased Natural Killer Cell and Increased Toll-Like Receptor/Inflammatory Gene Expression. J Neurotrauma. 2018;35(15): 1819-1829. [61] WANG Q, LIU L, CAO J, et al. Weighted gene co-expression network analysis reveals that CXCL10, IRF7, MX1, RSAD2, and STAT1 are related to the chronic stage of spinal cord injury. Ann Transl Med. 2021;9(15): 1248. [62] GE X, TANG P, RONG Y, et al. Exosomal miR-155 from M1-polarized macrophages promotes EndoMT and impairs mitochondrial function via activating NF-kappaB signaling pathway in vascular endothelial cells after traumatic spinal cord injury. Redox Biol. 2021;41:101932. [63] VAN BROECKHOVEN J, SOMMER D, DOOLEY D, et al. Macrophage phagocytosis after spinal cord injury: when friends become foes. Brain. 2021;144(10):2933-2945. [64] FAN H, TANG HB, SHAN LQ, et al. Quercetin prevents necroptosis of oligodendrocytes by inhibiting macrophages/microglia polarization to M1 phenotype after spinal cord injury in rats. J Neuroinflammation. 2019;16(1):206. [65] WANG Y, LI W, WANG M, et al. Quercetin prevents the ferroptosis of OPCs by inhibiting the Id2/transferrin pathway. Chem Biol Interact. 2023,381:110556. [66] ZHU K, ZHENG Z, ZHANG YY, et al. A comprehensive and systematic review of the potential neuroprotective effect of quercetin in rat models of spinal cord injury. Nutr Neurosci. 2024;27(8):857-869. [67] WIKLUND L, SHARMA A, MURESANU DF, et al. TiO(2)-Nanowired Delivery of Chinese Extract of Ginkgo biloba EGb-761 and Bilobalide BN-52021 Enhanced Neuroprotective Effects of Cerebrolysin Following Spinal Cord Injury at Cold Environment. Adv Neurobiol. 2023;32: 353-384. [68] LIU G, DENG B, HUO L, et al. Temporal profiling and validation of oxidative stress-related genes in spinal cord injury. Brain Res Bull. 2023;205:110832. [69] HE Z, ZHANG C, LIANG JX, et al. Targeting Mitochondrial Oxidative Stress: Potential Neuroprotective Therapy for Spinal Cord Injury. J Integr Neurosci. 2023;22(6):153. [70] SOHN HM, HWANG JY, RYU JH, et al. Simvastatin protects ischemic spinal cord injury from cell death and cytotoxicity through decreasing oxidative stress: in vitro primary cultured rat spinal cord model under oxygen and glucose deprivation-reoxygenation conditions. J Orthop Surg Res. 2017;12(1):36. [71] KAUR J, MOJUMDAR A. A mechanistic overview of spinal cord injury, oxidative DNA damage repair and neuroprotective therapies. Int J Neurosci. 2023;133(3):307-321. [72] WU AG, ZHOU XG, QIAO G, et al. Targeting microglial autophagic degradation in NLRP3 inflammasome-mediated neurodegenerative diseases. Ageing Res Rev. 2021;65:101202. [73] ROCA FJ, WHITWORTH LJ, REDMOND S, et al. TNF Induces Pathogenic Programmed Macrophage Necrosis in Tuberculosis through a Mitochondrial-Lysosomal-Endoplasmic Reticulum Circuit. Cell. 2019; 178(6) 1344-1361. [74] ZHANG S, ZHANG J, GUO D, et al. Biotoxic effects and gene expression regulation of urban PM(2.5) in southwestern China. Sci Total Environ. 2021;753:141774. [75] WU YR, CHANG KH, CHAO CY, et al. Association of SOD2 p.V16A polymorphism with Parkinson’s disease: A meta-analysis in Han Chinese. J Formos Med Assoc. 2021;120(1 Pt 2):501-507. [76] LIU W, ZHU J, WU Z, et al. Insight of novel biomarkers for papillary thyroid carcinoma through multiomics. Front Oncol. 2023;13:1269751. |
| [1] | Zhao Jiyu, Wang Shaowei. Forkhead box transcription factor O1 signaling pathway in bone metabolism [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1923-1930. |
| [2] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [3] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [4] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [5] | De Ji, Suo Langda, Wei Yuchen, Wang Bin, Awangcuoji, Renqingcuomu, Cui Jiuzeng, Zhang Lei, Ba Gui. Comprehensive analysis of genes related to endometrial receptivity and alternative splicing events in northwest Tibetan cashmere goats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1429-1436. |
| [6] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [7] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [8] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [9] | Zhao Ruihua, Chen Sixian, Guo Yang, Shi Lei, Wu Chengjie, Wu Mao, Yang Guanglu, Zhang Haoheng, Ma Yong. Wen-Shen-Tong-Du Decoction promoting spinal cord injury repair in mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1118-1126. |
| [10] | He Guanghui, Yuan Jie, Ke Yanqin, Qiu Xiaoting, Zhang Xiaoling. Hemin regulates mitochondrial pathway of oxidative stress in mouse chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1183-1191. |
| [11] | He Bo, Chen Wen, Ma Suilu, He Zhijun, Song Yuan, Li Jinpeng, Liu Tao, Wei Xiaotao, Wang Weiwei, Xie Jing . Pathogenesis and treatment progress of flap ischemia-reperfusion injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1230-1238. |
| [12] | Wang Rongrong, Huang Yushan, Li Xiangmiao, Bai Jinzhu. Prostaglandin E1 regulates vascular-related factors and protects microcirculatory function during the acute phase of traumatic spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 958-967. |
| [13] | Lu Ranran, Zhou Xu, Zhang Lijie, Yang Xinling. Dimethyl fumarate alleviates nerve damage in a mouse model of Parkinson’s disease [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 989-994. |
| [14] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [15] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||