Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (20): 4379-4388.doi: 10.12307/2025.687
Previous Articles Next Articles
Single-cell transcriptome analysis of mesenchymal and epithelial cells from four patients with polydactyly in the GEO public database
Fu Dongsheng1, 2, Aikeremujiang • Muheremu1, 2
- 1Department of Orthopedics, Sixth Affiliated Hospital of Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China; 2Key Laboratory of Orthopedic Regenerative Medicine, Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China
-
Received:2024-03-29Accepted:2024-08-27Online:2025-07-18Published:2024-12-25 -
Contact:Aikeremujiang • Muheremu, MD, Chief physician, Department of Orthopedics, Sixth Affiliated Hospital of Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China; Key Laboratory of Orthopedic Regenerative Medicine, Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China -
About author:Fu Dongsheng, Associate chief physician, Department of Orthopedics, Sixth Affiliated Hospital of Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China; Key Laboratory of Orthopedic Regenerative Medicine, Xinjiang Medical University, Urumqi 830001, Xinjiang Uygur Autonomous Region, China -
Supported by:the Natural Science Foundation of Xinjiang Uygur Autonomous Region, No. 2020D01C196 (to FDS)
CLC Number:
Cite this article
Fu Dongsheng, , Aikeremujiang • Muheremu, . Single-cell transcriptome analysis of mesenchymal and epithelial cells from four patients with polydactyly in the GEO public database[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(20): 4379-4388.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
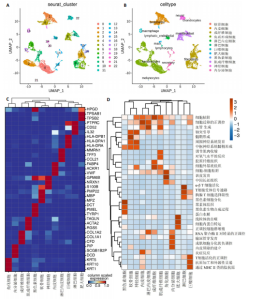
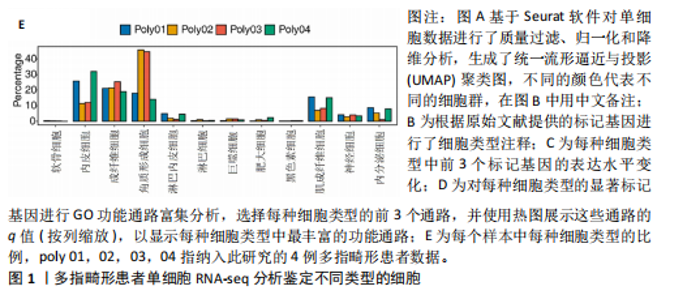
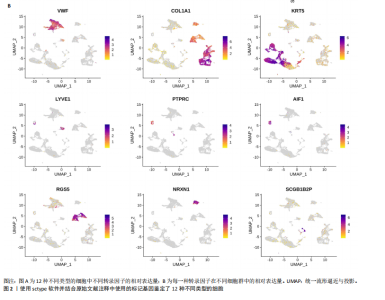
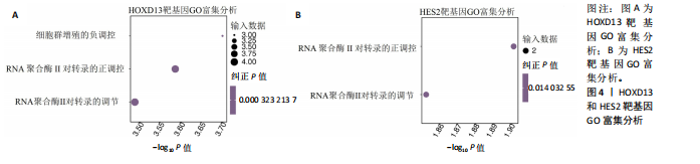
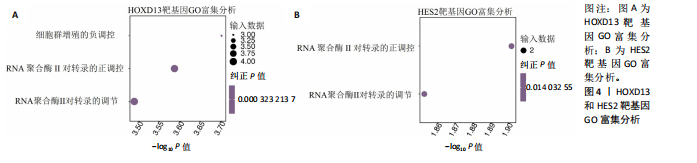
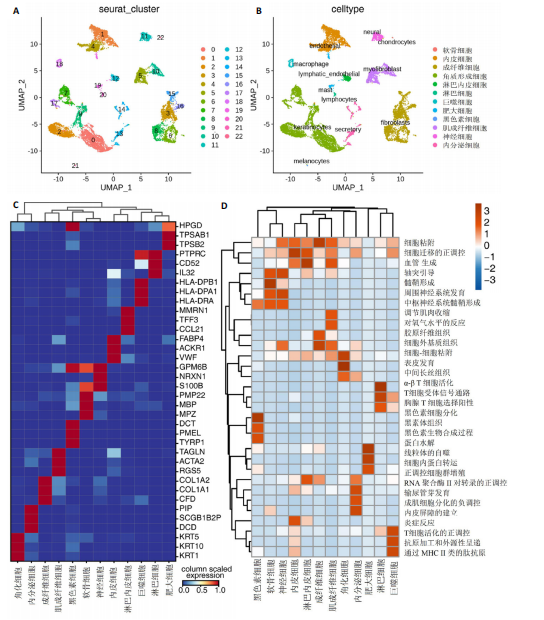
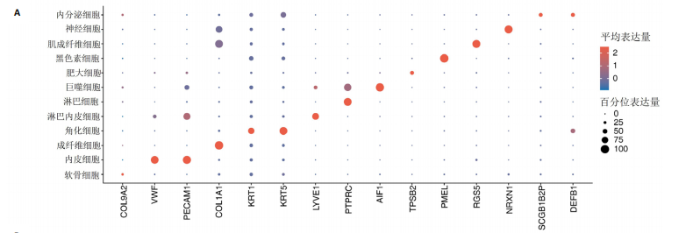
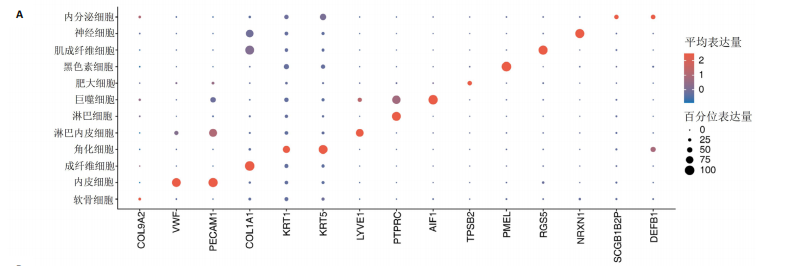
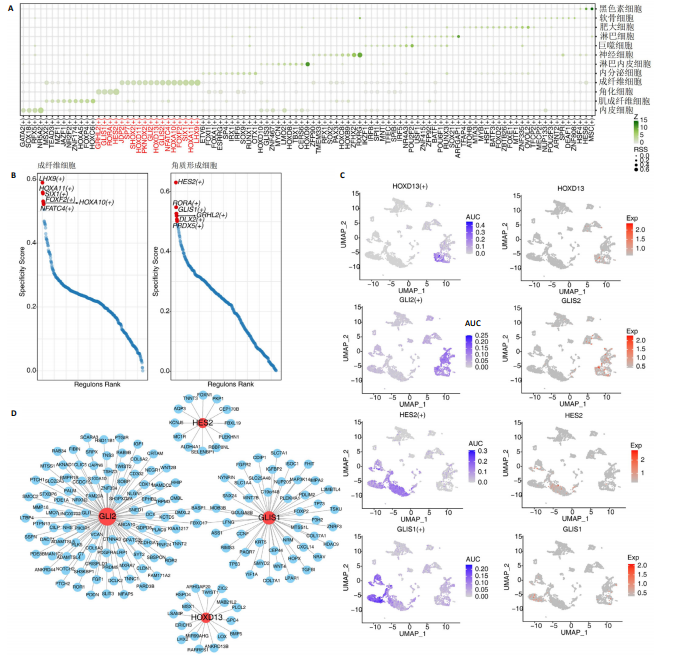
2.1 多指畸形患者单细胞RNA-seq分析鉴定不同类型的细胞 为了更深入地探索多指畸形发展中细胞特异性的关键调控因子,下载GSE158970多指畸形样本的数据,包括4例多指畸形患者的细胞间质和上皮细胞的单细胞数据。首先,在严格的数据质量控制后,获得了11 806个单细胞的转录谱数据。对这11 806个单细胞的转录表达矩阵进行了归一化处理和主成分分析,并选择前50个主成分进行统一流形逼近与投影(uniform manifold approximation and projection,UMAP)降维和可视化。经过无偏聚类分析,获得了23个细胞亚群(聚类)。使用sctype软件并结合原始文献注释中使用的标记基因,鉴定出了12个不同的细胞类型(图1A,B,图2)。同时,还显示了每个细胞类型的前3个标记基因的表达和富集功能通路(|avg_log2FC|≥0.5,P_val_adj≤0.05)(图1C,D)。发现不同细胞类型的标记基因和富集功能存在显著差异。例如,黑素细胞的显著标记基因主要与黑素细胞分化、黑素囊泡组织和黑色素生物合成相关,表明黑素细胞在黑色素分泌和皮肤色素沉着中可能起着重要作用。接下来,分析了每个样本中每种细胞类型的比例(图1E),发现主要的细胞类型包括成纤维细胞、角质细胞和内皮细胞。 2.2 单细胞分析揭示了与多指畸形细胞功能高度相关的调控因子 为了调查多指畸形不同细胞类型调控因子的调控情况,计算了各种细胞群体中转录因子的活性。分别鉴定了在成纤维细胞和角质细胞中特异激活的14个和4个调控因子。图3A,B分别显示了成纤维细胞和角质细胞相关调控因子的特异性得分。成纤维细胞中的调控因子HOXD13(+)和GLI2(+)在AUC评分上更高,并且这两个转录因子的表达在成纤维细胞中也更具特异性。角质细胞中的调控因子HES2(+)和GLIS1(+)在AUC评分上较高,这两个转录因子的表达在角质细胞中也相对特异(图3C)。 为了进一步验证这些转录因子的调控功能,构建了转录因子及其靶基因之间的调控网络(图3D)。其中,GLI2在成纤维细胞中调控的靶基因数量很多。GO富集分析显示在突触后膜组装、背腹极化图案形成和细胞迁移通路上富集(图3E),表明GLI2转录因子可能在多指畸形的形成过程中被激活。此外,HOXD13也被广泛报道与多指畸形的发生和发展密切相关,其突变可以导致人类和小鼠的肢体异常。HOXD13在指间间质中表达,并作为软骨形成和细胞分化的抑制因子。缺乏HOXD13会减少指融合处的厚度和长度,由此引起多指畸形,产生额外的、延长的手指在轴后面。角质细胞中HES2调控的靶基因主要与转录调控有关(图4),GLIS1调控的靶基因主要与增殖和凋亡通路相关(图3F),提示它们在维持细胞功能方面发挥作用。"
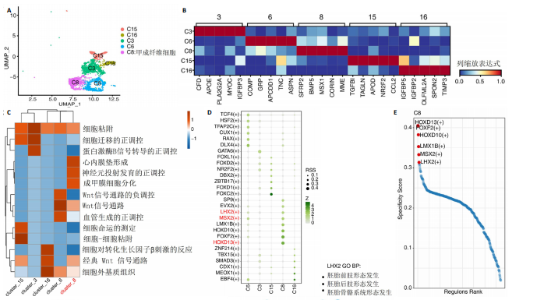
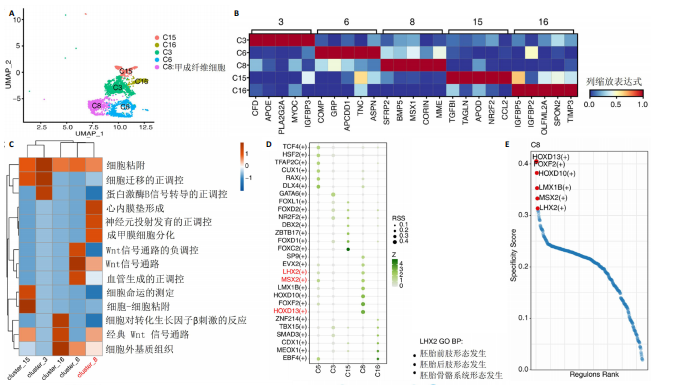
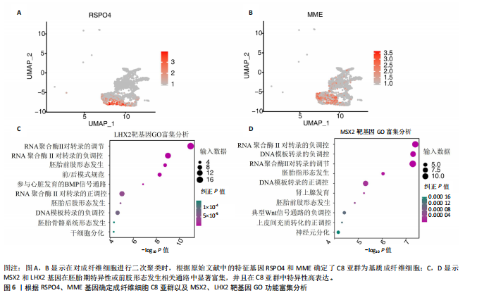
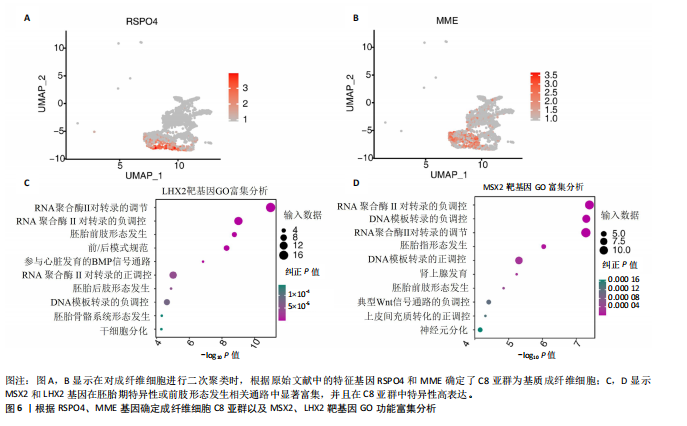
2.3 成纤维细胞中的转录因子活性与多指畸形 为了进一步研究不同成纤维细胞亚群中的转录因子活性,对成纤维细胞进行了二次聚类,共得到了5个亚群(图5A),其中C8基于原始文献中鉴定的特征基因RSPO4和MME被确定为甲基基质成纤维细胞(基质成纤维细胞)(图6A,B)。同时,发现每个细胞亚群中的标记基因表达具有明显的特异性(图5B)。此外,这些细胞亚群中的富集功能通路也显示出显著差异。例如,C8亚群主要与心内膜垫形成、神经发育和牙釉质母细胞分化相关(图5C)。此外,计算了每个细胞亚群中调控因子的特异性得分(图5D)。HOXD13、MSX2和LHX2在C8亚群中显著激活(图5E),它们的AUC活性和表达水平在C8亚"
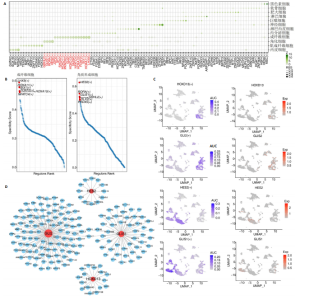
群中具有一定的特异性(图5F)。为了进一步验证这些转录因子的调控功能,构建了HOXD13、MSX2和LHX2转录因子及其靶基因的调控网络(图5G)。MSX2和LHX2的靶基因主要富集在胚胎期特异性或前肢形态发生相关的通路中,并且它们在C8亚群中特定性高表达(图5H,6C,D),提示C8亚群可能与多指畸形的发展有关联,但其关联性还需要通过探讨C8亚群在不同患者样本间及患病与正常样本中的比例差异、基质成纤维细胞与多指畸形发生发展相关性以及通过免疫组化或免疫荧光进一步证明。 2.4 多指畸形患者角质细胞中的转录因子活性 为了更全面地研究角质层细胞中各个细胞亚群的转录因子活性,进行了角质层细胞群的二次聚类,共得到5个亚群,包括基底角质细胞(Basal_kera)、表皮上皮细胞(Suprabasal_kera)、增殖角质细胞"
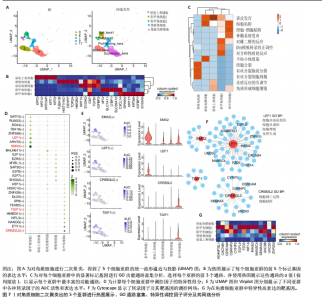
(Proliferating_kera)和两种类型的指甲角质细胞(Nail_kera1和Nail_kera2)(图7A),发现这些细胞亚群中标记基因表达和富集功能通路既有共性又有特异性(图7B,C)。为了进一步确定负责指甲形成的特异转录因子,计算了每个细胞亚群中调控因子的特异性得分(图7D)。其中,EMX2和LEF1在Nail_kera1亚群中显著激活,并且在Nail_kera1亚群中表现出较高的AUC活性和表达水平。此外,CREB3L2和TGIF1在Nail_kera2亚群中特异激活,并且在Nail_kera2亚群中表现出较高的AUC活性和表达水平(图7E)。为了进一步验证这些转录因子的调控功能,构建了EMX2、LEF1、CREB3L2和TGIF1与其靶基因的调控网络(图7F)。GO功能富集显示,LEF1的靶基因主要富集在细胞外基质、细胞增殖和血管生成等信号通路中,而CREB3L2的靶基因主要富集在凋亡和细胞黏附等过程中;此外,这些靶基因在Nail_kera细胞亚群中特异性高表达(图7G,图8)。基于这些结果,推测Nail_kera角质细胞亚群中的转录因子与指甲的形成和发展可能密切相关。"

| [1] AHMAD Z, LIAQAT R, PALANDER O, et al. Genetic overview of postaxial polydactyly: Updated classification. Clin Genet. 2023; 103(1):3-15. [2] BUBSHAIT DK. A review of polydactyly and its inheritance: Connecting the dots. Medicine (Baltimore). 2022;101(50): e32060. [3] KYRIAZIS Z, KOLLIA P, GRIVEA I, et al. Polydactyly: Clinical and molecular manifestations. World J Orthop. 2023;14(1): 13-22. [4] ÁLVAREZ LFG, TENORIO-CASTAÑO J, POLETTA FA, et al. A large, ten-generation family with autosomal dominant preaxial polydactyly/triphalangeal thumb: Historical, clinical, genealogical, and molecular studies. Am J Med Genet A. 2023;191(1):100-107. [5] IANEVSKI A, GIRI AK, AITTOKALLIO T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun. 2022;13(1):1246. [6] LEE J, HYEON DY, HWANG D. Single-cell multiomics: technologies and data analysis methods. Exp Mol Med. 2020;52(9):1428-1442. [7] VAN DE SANDE B, FLERIN C, DAVIE K, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15(7):2247-2276. [8] LIU S, WANG Z, ZHU R, et al. Three Differential Expression Analysis Methods for RNA Sequencing: limma, EdgeR, DESeq2. J Vis Exp. 2021;(175):e62528. [9] MA A, WANG C, CHANG Y, et al. IRIS3: integrated cell-type-specific regulon inference server from single-cell RNA-Seq. Nucleic Acids Res. 2020;48(W1): W275-W286. [10] BU D, LUO H, HUO P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317-W325. [11] MITSIS T, EFTHIMIADOU A, BACOPOULOU F, et al. Transcription factors and evolution: an integral part of gene expression. World Acad Sci J. 2020;2(1):3-8. [12] LOO CKC, PEAREN MA, RAMM GA. The Role of Sonic Hedgehog in Human Holoprosencephaly and Short-Rib Polydactyly Syndromes. Int J Mol Sci. 2021; 22(18):9854. [13] HAMELIN A, CONCHOU F, FUSELLIER M, et al. Genetic heterogeneity of polydactyly in Maine Coon cats. J Feline Med Surg.2020;22(12):1103-1113. [14] XU C, YANG X, ZHOU H, et al. A novel ZRS variant causes preaxial polydactyly type I by increased sonic hedgehog expression in the developing limb bud. Genet Med. 2020;22(1):189-198. [15] JIAO H, WALCZAK BE, LEE MS, et al. GATA6 regulates aging of human mesenchymal stem/stromal cells. Stem Cells. 2021;39(1): 62-77. [16] YIN J, GUO Y. HOXD13 promotes the malignant progression of colon cancer by upregulating PTPRN2. Cancer Med. 2021; 10(16):5524-5533. [17] DESALVO J, BAN Y, LI L, et al. ETV4 and ETV5 drive synovial sarcoma through cell cycle and DUX4 embryonic pathway control. J Clin Invest. 2021;131(13):e141908. [18] PAN J, FANG S, TIAN H, et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer. 2020;19(1):9. [19] TAN T, XU Z, GAO C, et al. Influence and interaction of resting state functional magnetic resonance and tryptophan hydroxylase-2 methylation on short-term antidepressant drug response. BMC Psychiatry. 2022;22(1):218. [20] PELED A, SARIG O, MOHAMAD J, et al. Dominant frontonasal dysplasia with ectodermal defects results from increased activity of ALX4. Am J Med Genet A. 2023; 191(12):2806-2812. [21] BUTLER A, HOFFMAN P, SMIBERT P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [22] RITCHIE ME, PHIPSON B, WU D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [23] IANEVSKI A, GIRI AK, AITTOKALLIO T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat Commun. 2022;13(1):1246. [24] AIBAR S, GONZÁLEZ-BLAS CB, MOERMAN T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083-1086. [25] SUO S, ZHU Q, SAADATPOUR A, et al. Revealing the Critical Regulators of Cell Identity in the Mouse Cell Atlas. Cell Rep. 2018;25(6):1436-1445.e3. [26] XIE C, MAO X, HUANG J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue): W316-W322. [27] JOVIC D, LIANG X, ZENG H, et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med. 2022;12(3):e694. [28] SLOVIN S, CARISSIMO A, PANARIELLO F, et al. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol Biol. 2021;2284:343-365. [29] MEREU E, LAFZI A, MOUTINHO C, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020;38(6):747-755. [30] KIM N, KIM HK, LEE K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020; 11(1):2285. [31] ZHANG Y, WANG D, PENG M, et al. Single‐cell RNA sequencing in cancerresearch. J Exp Clin Canc Res. 2021;40:81. [32] PAIK DT, CHO S, TIAN L, et al. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020;17(8):457-473. [33] LIAO J, YU Z, CHEN Y, et al. Single-cell RNA sequencing of human kidney. Sci Data. 2020;7(1):4. [34] WU Y, ZHANG K. Tools for the analysis of high-dimensional single-cell RNA sequencing data. Nat Rev Nephrol. 2020; 16(7):408-421. [35] KIM HJ, SHIM JH, PARK JH, et al. Single-cell RNA sequencing of human nail unit defines RSPO4 onychofibroblasts and SPINK6 nail epithelium. Commun Biol. 2021;4(1):692. |
| [1] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [2] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [3] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [4] | Aikepaer · Aierken, Chen Xiaotao, Wufanbieke · Baheti. Osteogenesis-induced exosomes derived from human periodontal ligament stem cells promote osteogenic differentiation of human periodontal ligament stem cells in an inflammatory microenvironment [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1388-1394. |
| [5] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [6] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [7] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [8] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [9] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [10] | Liu Lu, Zhong Chang, Yu Xin, Ren Chenyuan, Gong Yangyang, Zhou Ping, Wang Yingbin. Academic progress and clinical application of in vitro synthetic microenvironment to promote maturation of human pluripotent stem cell-derived cardiomyocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7856-7862. |
| [11] | Li Zhongzheng, Chen Zhenghao, Tang Ziyou, Lou Kaiyang, Zhang Rui, Liu Qi, Zhao Na, Yang Kun. Effects of scaffold materials combined with biological factors on biological characteristics of dental follicle cell proliferation and osteogenic differentiation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(34): 7405-7414. |
| [12] | Tian Yushi, Fu Qiang, Li Ji . Bioinformatics identification and validation of mitochondrial genes related to acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6697-6707. |
| [13] | Zhang Pulian, Liu Baoru, Yang Min . Mesenchymal stem cells for treatment of aplastic anemia: inhibiting or activating relevant targets in its pathological evolution [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6800-6810. |
| [14] | Wang Jianxu, Dong Zihao, Huang Zishuai, Li Siying, Yang Guang. Interaction between immune microenvironment and bone aging and treatment strategies [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6509-6519. |
| [15] | Zhu Xuekun, Liu Heng, Feng Hui, Gao Yunlong, Wen Lei, Cai Xiaosong, Zhao Ben, Zhong Min. Identification of core genes of osteoarthritis by bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(3): 637-644. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||