Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (13): 2822-2831.doi: 10.12307/2025.044
Previous Articles Next Articles
Application of single-cell RNA sequencing in spinal cord injury
Wang Peigeng1, Ye Dongping1, 2
- 1Guizhou Medical University, Guiyang 550025, Guizhou Province, China; 2Guangzhou Red Cross Hospital, Guangzhou 510220, Guangdong Province, China
-
Received:2023-10-25Accepted:2024-03-09Online:2025-05-08Published:2024-09-12 -
Contact:Ye Dongping, MD, Chief physician, Master’s supervisor, Guizhou Medical University, Guiyang 550025, Guizhou Province, China; Guangzhou Red Cross Hospital, Guangzhou 510220, Guangdong Province, China -
About author:Wang Peigeng, Master candidate, Guizhou Medical University, Guiyang 550025, Guizhou Province, China -
Supported by:Guangzhou Science and Technology Planning Project, No. 202102010111 (to YDP)
CLC Number:
Cite this article
Wang Peigeng, Ye Dongping. Application of single-cell RNA sequencing in spinal cord injury[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(13): 2822-2831.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

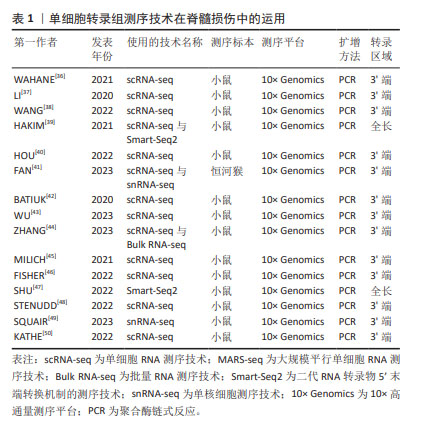
2.1 单细胞转录组测序技术的研究历程 随着单细胞转录组测序技术的进步,研究人员可以在单细胞水平上描绘出组织的异质性。自从TANG等[14]在2009年首次报道单细胞转录组测序技术后,在过去的近15年里,许多更灵敏、更准确的单细胞转录组测序技术被开发出来。其中,Quartz-Seq 技术对TANG等[14]的方法进行了优化,简化了实验流程[15]。CEL-seq技术于 2012年用体外转录代替了PCR[16],2014 年开发的MARS-seq技术与CEL-seq技术很类似[17]。2012美国和瑞典的科学家共同开发了Smart-Seq技术,SMART 技术使Smart-Seq和Smart-Seq2技术成为目前应用较多的技术[18-19],Smart-seq3技术则于2020年由瑞典卡罗林斯卡学院的Rickard团队进一步改进并发表[20]。后来,Drop-seq和inDrop两种技术在哈佛大学问世,让分析单细胞的基因表达成为现实[21-22]。单细胞测序技术的丰富与完善,揭示了单个细胞的基因结构和基因表达状态,反映细胞间的异质性。该技术为复杂和动态系统的细胞组成提供了前所未有的视角,特别是单细胞转录组学方法已经引领了技术进步,使细胞表型的绘制更加全面[23]。目前,在肿瘤学、微生物学、神经病学、生殖学、免疫学、消化系统和泌尿系统等领域发挥重要作用[24-26],已正成为生命科学研究的焦点。单细胞转录组测序技术的主要发展历程见图3。"
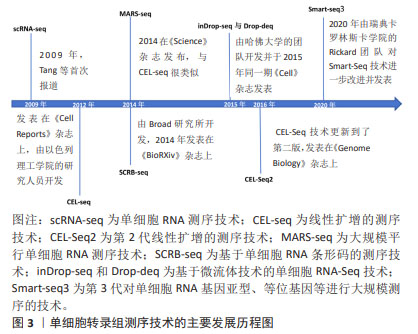
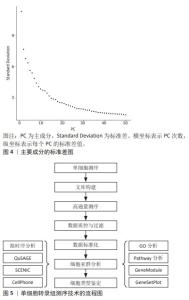
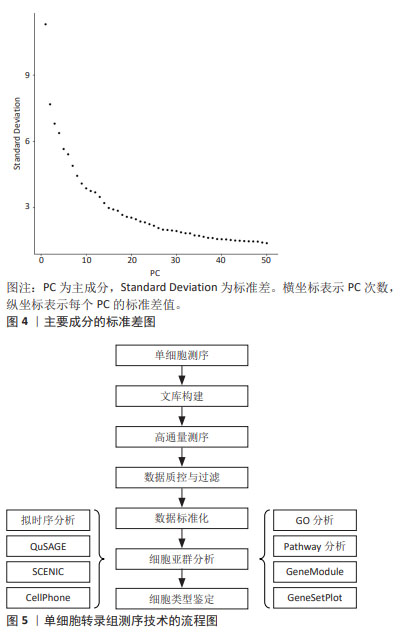
2.2 主要的单细胞转录组测序方法介绍 单细胞转录组测序技术的方法主要可分为13种,包括CEL-Seq2,MARS-Seq,Quartz-Seq2,gmcSCRB-Seq,Smart-Seq2,ddSEQ,ICELL8,C1HT-Small,C1HT-Medium,Chromium,Chromium (sn),Drop-seq以及inDrop。MEREU等[27]对这常用的13种单细胞RNA测序方法进行多方面的性能评估,最终研究发现Quartz-Seq2方法在众多方案中最为准确,紧随其后的是CEL-Seq2和Chromium。其中,单细胞RNA测序技术是运用最多、最广泛的,Smart-Seq2技术的基因转录区域多为全长段,常与单细胞转录组测序技术结合使用。令人遗憾的是Quartz-Seq2技术在脊髓损伤的研究尚未有过相关报道。 2.3 单细胞转录组测序技术步骤 2.3.1 样品的制备与单细胞分离 针对单个细胞研究时,首先将单个细胞进行分离,并确保其生物完整性不被破坏。通常,从细胞群体中分离出研究所需的单细胞,有多种方法可供选择,包括连续稀释法、显微操作法、激光捕获显微切割术、拉曼镊子技术、荧光激活细胞分选术和微流控技术等[28]。研究者可根据通量的要求,以及是盲选还是有偏向的选择,对这些要求选择单细胞分离选取。在样品制备过程中,优化样本获取和组织加工条件对于避免可能限制细胞图谱价值的成分偏差和基因表达伪因素至关重要[29]。因此,需要进行专门的研究,以确定在健康和疾病背景下组织和器官制备的最佳条件。 2.3.2 细胞溶解与基因组获取 对细胞进行溶解来获取基因组(DNA或RNA),这步骤非常关键,应尽量保证基因组的完整性。目前细胞溶解的方法可以分为3大类:物理法、化学法和生物酶降解法。 2.3.3 全基因组扩增 多步骤的转录组扩增,包括反转录、转化为可扩增的cDNA和扩增[30]。基于测序仪的检测线,目前大多是利用 DNA 聚合酶和不同形式的引物来进行扩增的。最近,SHENG等[31]发现MATQ-seq技术比Smart-seq2技术在全基因组扩增上更加灵敏。而且,MATQ-seq技术提供了整个基因体的覆盖范围,并允许检测总RNA,这是一个重大进步。此外,ZONG等[32]还为单个细胞的基因组扩增而开发了一种名为MALBAC-RNA方法,该方法使用准线性而不是指数扩增来扩增和测序单个细胞的转录组,从而避免了扩增偏差。 2.3.4 数据标准化与测序文库的制备 进行数据分析时,需要对原始数据标准化为特定的格式,并根据特异分子标签,将拆解后的测序数据构建成单细胞基因表达矩阵用于下游分析。目前,10X Genomics和BD Rhapsody是最主流的2种高通量测序平台,也可在Illumina平台中测序,使用基于转座酶Tn5的片段进行文库制备[33]。下游数据结合Scran,Seurat和Scater等软件包的分析,降低了因测序深度、表达模式不同而导致的偏倚,提高数据的可信度[34]。 2.3.5 降维和可视化 标准化后,可以使用无偏聚类算法来根据基因表达谱确定哪些细胞密切相关。主成分分析通常是选择的聚类算法,可以预测多维数据的相关性。成分分析将相关基因的信息合并为一个“元基因”或主成分(Principal componet,PC)决定,PC的排名越低,它对解释数据方差所起的作用就越小。使用排名较低的PC通常是不利的,因为它增加了计算负载。因此,决定使用多少台PC进行可视化非常重要[35]。如图4所示,绘制每个PC的标准差以表示该PC中包含的方差,PC1表示该数据具有最大方差,而且标准差最大;PC2表示该数据具有第二大的方差,依此类推。根据解释数据方差的重要性对PC进行排名,PC排名越低,对解释数据方差贡献越小。图4中显示了PC10附近的弯头,表示在第10次PC之后,对方差解释的贡献趋于平稳,成为可视化中PC包含的截止点。T-分布式随机邻居嵌入是一种常见的可视化方法。基因水平分析主要用于基因的富集分析、调控网络以及寻找差异表达基因等方向,而细胞水平分析主要体现在聚类分析和轨迹推断。单细胞转录组测序技术的具体流程图如图5所示。"

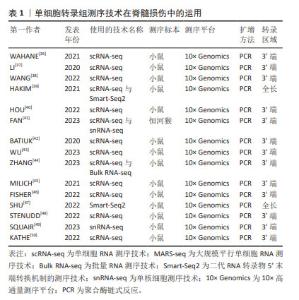
2.4.2 胶质细胞的异质性 小胶质细胞:小胶质细胞可快速协调损伤反应,WAHANE等[36]结合了骨髓特异性转录组学和单细胞转录组测序技术,发现了4种不同类型的小胶质细胞,包括免疫性MG1群体、反应性MG2群体、立即反应性MG3群体和增殖性MG4群体,结果显示MG1主要承担免疫相关功能,表达更高水平的细胞因子(白细胞介素1a,白细胞介素1b和Cxcl-10)和免疫应答基因(肿瘤坏死因子,Ccl3和Nfkbia),并接合核转录因子kB和肿瘤坏死因子信号通路。MG2专门从事吞噬作用和脂质代谢,表达自噬和溶酶体基因(Ctsd,Ctsl,CD81,CD68和Tyrobp),并参与Tyrob网络和转化生长因子信号通路。MG3由脑源性神经营养因子信号传导和IEGs-Fos,Jun和Egr1的诱导来定义,揭示了更本质的反应性质。MG4表示增殖状态,并被增殖基因(Cdk1,Top2a,Nusap1和Humg2)和p53信号标记。在脊髓损伤后,连续的细胞轨迹从MG3群体的激活开始,发展到免疫聚焦的MG1群体和反应性MG2群体,然后发展到增殖性MG4群体。由此可以看出,小胶质细胞在脊髓损伤后立即发出神经营养信号,再结合其免疫功能和增殖功能一起对损伤的脊髓做出快速反应,以达到修复脊髓损伤的目的。该研究不仅揭示了小胶质细胞的神经信号传导功能、免疫功能和增殖功能,还在时间的不同基因程序上反映了小胶质细胞的异质性。遗憾的是,该研究未进行额外的免疫染色和功能实验验证其研究成果,还缺乏临床模型与体内外实验。 LI等[37]对脊髓挤压伤的新生小鼠进行了单细胞转录组测序,将得到的所有小胶质细胞族群重新聚集成5个转录不同的细胞簇,命名为MG0-MG4。特征基因为P2ry12,Tmem119和Siglech的MG0被认为与完整脊髓中的小胶质细胞稳态有关;特征基因为CD9和Serpine2的MG1被认为与典型的小胶质细胞活化有关;MG2的特征基因为Fn1,MG2被认为是MG1和MG0之间的中间状态;MG3的特征基因为Ms4a7,Ms4a6c和MG3在细胞外基质、伤口愈合、吞噬作用、血管生成以及免疫反应和内肽酶的负调控中功能显著富集,这意味着MG3簇可能具有促进无瘢痕愈合的能力。MG3和MG1细胞表现出互补分布,MG3细胞直接位于病变中和病变周围,MG1细胞位于病变周围的脊髓中,而且MG3和MG1细胞都表达典型的小胶质细胞激活标记物,如Spp1和Igf1。此外,MG3簇群在新生儿小胶质细胞中分泌纤连蛋白和结合蛋白,并形成了细胞外基质的连接,连接脊髓的断端。MG3还表达几种细胞外和细胞内肽酶抑制剂,如Cstb,Stfa1和Serpinb6a,以及促分解介质Anxa1,这可能有助于它们快速分解炎症,改善小胶质细胞的愈合和轴突再生。因此,在脊髓损伤的急性期,促愈合的小胶质细胞发挥着多种作用,不仅在段端之间形成了有效的桥接,而且表现出了显著的炎症消退及促愈合特性。此外,与成人小胶质细胞相比,新生儿损伤将稳态小胶质细胞转化为具有激活和去分化特征的状态,并且保持永久活跃,这对于无瘢痕伤口愈合至关重要。虽然该研究指出了新生小鼠通过自发的轴突再生开始无瘢痕损伤的愈合,但下行轴突或椎管内重组的作用尚不清楚。因此,未来的研究重点应该在恢复稳态小胶质细胞的下游轴突信号和椎管内的重组作用机制。 WANG等[38]使用单细胞转录组测序技术描绘了小鼠脊髓损伤早期和晚期免疫细胞的动态转录景观,发现了小胶质细胞被分为4类型(7个簇),其中包含干扰素反应小胶质细胞、稳态小胶质细胞、3个损伤相关小胶质细胞簇和2个增殖相关小胶质簇。其中,损伤相关小胶质细胞簇富集了与干扰素反应、细胞因子产生、脂质代谢、胶质生成、神经胶质细胞迁移、成纤维细胞增殖和抗原呈递相关的基因本体术语,提示损伤相关小胶质细胞簇有助于组织修复。干扰素反应小胶质细胞和稳态小胶质细胞主要出现在健康的脊髓中,并且依赖于性别分布。增殖相关小胶质瘤簇与氧化应激和增殖都有关。与前面描述的研究相似,HAKIM等[39]从雌性小鼠创伤性脊髓损伤中心采集了免疫细胞的时间动力学。该研究发现了一种标志物为Myc,Glmp,Nfe2l2和Skil的小胶质细胞簇,该亚簇通过活性细胞状态从永久重新编程的基线小胶质细胞进化而来。这些细胞在脊髓严重损伤后几天出现,并在损伤部位的人类脊髓中保留多年;最后,小胶质细胞耗竭实验表明,该亚簇与在神经变性或脱髓鞘中观察到的亚簇类似,此类小胶质细胞的激活对脊髓损伤的恢复有很强的积极作用。 在以上的研究中发现,脊髓损伤后的小胶质细胞具备增殖、免疫和修复等功能,已被证明在协调脊髓损伤反应中起着至关重要的作用。在未来的研究中,从小胶质细胞的异质性出发,确定参与恢复稳态小胶质细胞的其他分子组成值得进一步期待。 星形胶质细胞:最新的一项研究表明,星形胶质细胞谱系对损伤的微环境变得具有反应性,它们在神经保护和修复中也起着至关重要的作用[51]。星形胶质细胞目前被归类为A1和A2两种类型,神经炎症诱导的A1型星形胶质细胞上调了许多对突触具有破坏性的基因,被认为是有害的;相反,缺血诱导的A2型星形胶质细胞下调了许多促进神经元存活和生长的神经营养因子,表明A2型星形胶质是有利的[52]。A1型特异性表达Rela,干扰素γ,C1q,Lcn2,H2-T23,Serping1,H2-D1,白细胞介素1β,肿瘤坏死因子和补体C3;A2型特异性表达脑源性神经营养因子,Il6,Emp1,S100a10,Slc10a6,Lif,Clcf1,Thbs1/2和Tm4sf1[53]。HOU等[40]在小鼠半切脊髓后构建了许多星形胶质细胞的相关亚群,其中簇3、簇4和簇6在脊髓损伤后的急性和亚急性时间表达A2型基因,例如S100a10,Thbs1和Tm4sf1,在脊髓损伤后的中期,簇3和簇4高度表达C1q和H2-D1的A1型基因。簇1、簇2、簇5、簇9在脊髓损伤后的急性期也表达Thbs1,被定义为A2型星形胶质细胞。然后,簇1、簇2、簇5、簇9在脊髓损伤急性期高表达Lcn2和H2-D1,因此这些细胞被鉴定为A1型星形胶质细胞。高表达Cryab的簇7和簇11细胞被认为是A2型星形胶质细胞。其中,Cryab已被证明通过抑制胱天蛋白酶活性抑制神经炎症并保护细胞免于凋亡,簇1和簇2、簇5、簇9细胞执行神经毒性细胞碎片和细胞毒性的清除。这些数据不仅明确了星形胶质细胞A1和A2两个类型的标记基因,而且有助于开发新的星形胶质细胞亚型作为脊髓损伤修复的靶点。尽管该研究已经分析了这些星形胶质细胞在每个簇中的功能,但未进一步使用遗传方法或其他适当的工具直接检测这些星形胶质族群的功能。 FAN等[41]利用更高级别的实验模型(恒河猴)作为研究对象,在进一步的星形胶质细胞聚类分析揭示了8种亚型,其中3个是损伤后新出现的,这些簇显示出不同的表达,指向白质或灰质星形胶质细胞;一个表达细胞周期基因MKI67和TOP2A的亚群被鉴定为分裂型星形胶质细胞。此外,一些被标记为激活的白质和灰质星形胶质细胞在损伤后的各个阶段都保持较高水平,而且在脊髓损伤后瘢痕形成和血管生成中发挥重要作用。一些星形胶质细胞被发现具有类似于人类胚胎脊髓神经前体细胞的基因,此发现突出了白质星形胶质细胞在损伤后的潜在可塑性。最后,该研究还发现脊髓损伤后星形胶质细胞的特定基因表达上调,例如SERPINA3,CHI3L1,胶质纤维酸性蛋白,PAX3,ASCL1和ITPRID1等。该研究在实验模型上做出了更高级别的尝试,其突破性值得肯定,但所研究的样本(恒河猴)使用量相对过少,缺乏体内外实验进一步证明。 GAO等[54]结合以往研究的成果,综述了星形胶质细胞在中枢神经系统发挥的控制和调节作用。高表达的Hes5,Ubc和Nrarp基因的星形胶质细胞亚群1(AC1)被认为与Notch信号通路密切相关,当Notch信号被阻断时,星形胶质细胞可被促进转化为神经干细胞样状态。高表达Btg2的亚群2(AC2)被认为是神经元生成祖先的标志,当Rbpj-k的缺失可诱导转录因子的激活,促进大脑皮质发育过程中的神经元特异性表达。此外,单个细胞运动轨迹的重建,确定了星形胶质细胞的神经发生和反应性胶质细胞的增生。该研究概述了星形胶质细胞在不同中枢神经系统疾病中的关键作用,使研究者们能够识别来自原生星形胶质细胞的皮质星形胶质细胞和神经祖细胞群。 BATIUK等[42]使用了单细胞转录组测序技术确定了成年小鼠皮质和海马体中的5个星形胶质细胞亚型,命名为AST1-AST5。其中,AST1高表达Gfap和Agt,主要存在于基底膜和海马体;AST2高表达Slc7a10,Gria2,Slitrk2和Sema4b,参与谷氨酸能的传递和突触的生长;AST3高表达Gabrg1,Etv5和Spon1,参与GABA能的传递和突触的生长;AST4高表达Cdk4,Sirt2,Ascl1,Emx1和Dab1,与有丝分裂、细胞周期、转录调节以及神经发生和神经元分化有关;AST5高表达Sirt2,Sept2和Emp2,与经典星形胶质细胞的葡萄糖代谢和能量产生密切相关。随后,该研究进一步证实了AST1和AST4主要是海马,AST2主要是皮层,AST3和AST5均匀分布在大脑区域之间。 大量研究表明星形胶质细胞不仅可划分为A1(不利)和A2(有利)两个类型,还可以分为炎性星形胶质细胞、神经保护性星形胶质细胞、神经元样星形胶质细胞和可塑性白质星形胶质细胞等多种类型。在脊髓损伤的病理机制上,星形胶质细胞以激活的方式在脊髓损伤中发挥许多重要功能,包括清除坏死组织、形成保护屏障、维持微环境平衡、与免疫细胞相互作用以及形成胶质瘢痕等。 少突胶质细胞:该细胞在其成熟阶段表现出不同的反应,FAN等[41]确定了2个新出现的与疾病相关的少突胶质细胞亚群,命名为DOL1和DOL2,与其他少突胶质细胞相比,它们高表达的基因为SERPINA3,NRXN3,MT2A和UBASH3B。其中,SERPINA3已被报道为神经退行性和自身免疫性炎症条件下疾病相关少突胶质细胞的关键标志物,DOL1和DOL2上调的基因共享许多生物学过程,如突触组织和组织迁移。然而,DOL1表达更多富含血管生成调节的基因,DOL2有更多与脂代谢相关的基因,而且DOL2表达更多对干扰素γ有反应的基因。值得注意的是,两个DOL簇均下调了髓鞘相关基因,提示髓鞘化基因表达减少。该研究还发现损伤后轴突及其髓鞘的变性触发小胶质细胞和其他细胞反应的激活,揭示了少突胶质前体细胞在脊髓损伤后一直被激活,但在腰椎区的反应较轻。 WU等[43]使用单细胞转录组学的方法注释了少突胶质细胞族群的主要标记基因为Gjb1,Ermn,Mog,Tmem125和Hapln2等。分化诱导的少突胶质细胞体在急性损伤阶段起着至关重要的作用,拟时序分析显示了少突胶质细胞谱系细胞基因表达的动态变化,构建的动态基因可以分为上调型、下调型和随着细胞周期波动型。其中,上调表达的基因倾向于表达成熟少突胶质细胞的基因,与髓鞘生成、轴突髓鞘形成、神经胶质细胞分化和胶质形成有关;波动表达的基因主要与Toll样受体4结合、RAGE受体结合、花生四烯酸结合和icosanoid结合,这反映了脊髓损伤后急性炎症反应的生物学过程。 长期以来,人们已经确定少突胶质细胞和少突胶质前体细胞在髓鞘的形成中起着关键作用。有学者认为脊髓损伤后,中枢区域出现广泛的少突胶质细胞死亡和长期脱髓鞘[55]。目前的研究显示脊髓损伤早期少突胶质细胞的大量缺失可能是神经功能严重受损的基础,而晚期少突胶质的缺失可能会进一步促进轴突损伤,导致神经功能更难恢复。然而,基于单细胞转录组测序技术研究少突胶质细胞及其更深层次的机制相对匮乏。因此,少突胶质细胞谱系细胞的异质性和细胞间相互作用值得进一步探索。表2列举了单细胞转录组测序技术在脊髓损伤的重点研究进展。"
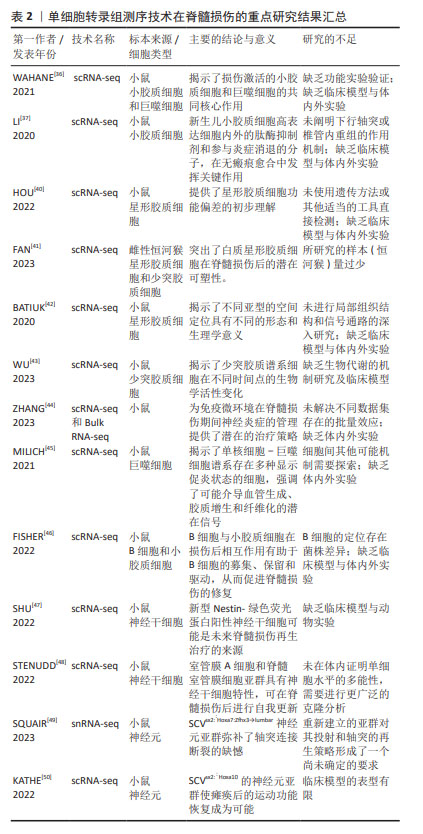

2.4.3 脊髓损伤后的免疫细胞浸润 免疫细胞在脊髓损伤后起着至关重要的作用。然而,免疫对脊髓损伤和修复的不完全了解阻碍了脊髓损伤疗法的发展。ZHANG等[44]基于单细胞转录组测序技术和Bulk RNA-seq探讨了脊髓损伤亚急性期的免疫枢纽基因和免疫细胞组成,该研究发现中性粒细胞、巨噬细胞、小胶质细胞、B淋巴细胞和T淋巴细胞都参与了脊髓损伤后的炎症反应。同时,免疫细胞的组成和表型也随着损伤的阶段和损伤微环境中的信号而改变。例如,小胶质细胞/巨噬细胞、T细胞和B细胞能够在受损脊髓中产生促炎和抗炎反应,从而调节脊髓损伤后的继发性损伤。一些获得性免疫细胞(静息树突状细胞、浆细胞和活化的NK细胞)的免疫微环境发生了变化,这与血脊髓屏障的破坏密切相关。重要的是,该研究表明调节免疫微环境的双重作用是管理神经炎症的潜在策略。SHANG等[56]发现脊髓损伤后中枢神经系统的天然免疫监视功能被激活,导致免疫细胞大量浸润。该分析在发现了免疫细胞浸润的2种亚型,C1簇群高表达PTPRC,LDHA,JAK2和B2M,被认为与卵泡辅助T细胞、调节性T细胞、静息NK细胞、单核细胞、M0巨噬细胞、静息树突状细胞和活化肥大细胞有关;C2簇群高表达TNFSF4,LAG3,JAK1,IL12B,IL12A,IFNG,ICOS和CD40LG,被认为与CD4记忆T细胞、浆细胞和活化肥大细胞有关。 巨噬细胞:与小胶质细胞类似,巨噬细胞对脊髓修复至关重要。MILICH等[45]使用单细胞RNA测序来评估几乎所有构成小鼠脊髓损伤部位的细胞类型,该研究在外周来源的髓系簇发现了2种亚型的巨噬细胞,即趋化性诱导巨噬细胞和炎性巨噬细胞。趋化性诱导巨噬细胞富含与其他白细胞(如中性粒细胞和粒细胞)趋化相关的基因本体术语;炎症性巨噬细胞的基因本体术语与胶质细胞和巨噬细胞的激活以及炎症反应有关。对这2种巨噬细胞亚型的进一步数据分析,揭示了单核细胞-巨噬细胞谱系中存在多种细胞状态,这些状态显示了促炎状态的时间进展。此外,通过评估配体-受体对在不同细胞类型上的表达,该研究强调了可能介导血管生成、胶质增生和纤维化的潜在信号关系。WAHANE等[36]也鉴定了4个巨噬细胞亚群,命名为Mac1-4。Mac1与细胞增殖、迁移和细胞外基质相关,参与了血管生成、脂质代谢和吞噬作用;而Mac2与M1巨噬细胞活化有关,调节细胞代谢(线粒体呼吸链复合体)和RNA过程(核糖体和剪接体);Mac3与前列腺素合成和晚期糖基化终产物活化的受体有关,涉及神经元相互作用,比如谷氨酸信号传导和神经系统发育;Mac4从事细胞外基质的相互作用,并参与了神经系统的发育过程和Hippo-Merlin信号传导。由于吞噬基因、抗炎标志基因、细胞外基质基因和营养因子的高表达,提示该数据集支持巨噬细胞的持久修复功能。 有关参与脊髓损伤的免疫细胞,特别是巨噬细胞的研究仍处于初始阶段。但目前的研究表明,当吞噬型巨噬细胞低表达促炎基因时,炎症型巨噬细胞则表达促炎和抗炎两类基因。此外,巨噬细胞在脊髓损伤后表现出较强的血管生成、脂质代谢和吞噬功能。脊髓损伤后神经元和轴突能否再生不仅取决于其原始生长能力,还取决于神经血管微环境和表观遗传学变化,主要体现在DNA甲基化和组蛋白乙酰化,这些新的见解可以帮助了解脊髓损伤病理生理学的复杂过程。 B细胞:FISHER等[46]通过对B细胞的聚类分析,在亚急性和慢性脊髓损伤的基因表达变化研究中得到了4个亚簇,命名为簇A-簇D。其中,簇A细胞表达B细胞受体基因;簇B细胞表达MHC-Ⅱ相关成分;簇C细胞表达高水平的IgM和IgG2b。而且,该研究发现B细胞处于多种发育和激活状态,B细胞与小胶质细胞在损伤后相互作用有助于B细胞的募集、保留和驱动,从而促进脊髓损伤的修复。 2.4.4 神经干细胞与神经元的异质性 脊髓损伤后的神经干细胞具有很大的修复潜力。SHU等[47]通过单细胞RNA测序技术对脊髓损伤前后的细胞进行表征,确定了神经干细胞的两个亚组,即位于中央管内的神经干细胞亚组和位于中央管外的神经干细胞亚组。位于中央管内的亚组脊髓损伤后不能启动神经干细胞的活性,而位于中央管外的亚组在脊髓损伤后被激活并表现出活跃的神经干细胞特性。最终,该研究还发现了源自中央管外的新型Nestin-GFP+神经干细胞。在完整的脊髓中,这些细胞是同质的,但它们在脊髓损伤后5 d被激活,因而具有增殖和分化能力。因此,与中央管内亚组的静止特征不同,中央管外的神经干细胞亚组可显示出显著的神经干细胞活性,并且在脊髓损伤后具有高度异质性,这表明它们可能是未来脊髓损伤再生治疗的来源。 STENUDD等[48]的研究结果显示室管膜细胞基因表达程序丧失,因为室管膜A细胞获得信号并在损伤后去分化为干细胞样的转录状态。该研究进一步阐明了高分化的室管膜A细胞和相对罕见的脊髓室管膜细胞亚群具有神经干细胞特性,能够在脊髓损伤后进行自我更新,这为脊髓修复的再生治疗干预提供了新的见解,但是,该研究未在体内证明单细胞水平的多能性,而且需要进行更广泛的克隆分析,在脊髓损伤后等待了更长的时间。 长期以来,科学家们注意到一个关于脊髓自然修复的有趣现象。人和实验动物在发生脊髓损伤后的头几个月,会出现不同程度的自发性功能恢复,而这种自愈现象与中胸段脊髓的神经元有关。为了研究神经元在脊髓损伤中的异质性,SQUAIR等[49]使用荧光蛋白标记和单细胞核RNA测序技术来鉴定了脊髓损伤后参与运动功能修复的神经元亚群。他们发现,在脊髓受损后恢复运动能力的小鼠中,有一类神经元的树突棘形态发生、突触增强程序和肌动蛋白细胞骨架重组的相关基因表达上调,该亚群高表达Vsx2的长距离投射神经元和Zfhx3 的Z群神经元,因此这类神经元被命名为SCV sx2::Hoxa7:Zfhx3→lumbar,简称SC神经元。进一步研究表明,在中胸段脊髓的不同神经元亚群中,SC神经元不仅是脊髓自然恢复过程中转录扰动最大的神经元亚群,而且还表现出相关的解剖学特征,使得来自脊髓上游的神经信号通过脊髓断裂面传递到腰椎脊髓中的运动神经元。这些研究结果不仅暴露了必须再生的特定神经元轴突,还揭示了这些轴突必须重新连接到它们的天然投射目标区域,才能真正修复脊髓损伤并恢复运动功能。这弥补了ANDERSON团队[57]因新生的轴突无法连接到脊髓断裂面另一侧导致小鼠的运动能力没有恢复的缺憾。更重要的是,基于上述发现,研究团队进一步开发了基因疗法用于治疗完全性脊髓损伤的小鼠,成功地大幅度恢复了小鼠瘫痪后肢的行走能力,这一突破性结果为脊髓损伤修复领域再创里程碑。 KATHE等[50]将单核 RNA 测序技术应用于这些小鼠的脊髓,发现了表达Vsx2和尾侧脊髓神经元标志物Hoxa10的神经元亚群,该类稳健的神经元亚群被命名为SCV sx2::Hoxa10。尽管这些神经元在脊髓损伤前不需要行走,但它们对于脊髓损伤后硬膜外电刺激的恢复至关重要,增强这些神经元的活动可以逐步实现步行的恢复,而消融它们则阻止了中度脊髓损伤后自发的步行恢复。此外,该研究结合单核 RNA 测序和空间转录组学建立了一个框架,用于使用分子制图来识别产生复杂行为的神经元,最终揭示了SCV sx2::Hoxa10神经元是瘫痪后行走恢复的基本要求。 总之,了解每个神经元亚群对行走等复杂行为的贡献是神经科学的一项挑战,有很多学者不仅找到了促进完全脊髓损伤恢复的神经元亚群,还证实促进此类神经元再生到自然靶区,可以让完全脊髓损伤小鼠的运动功能有实质性的恢复,这些结论指导了研究者们对运动恢复基础回路的研究。虽然这些研究结果在应用于人类之前,必须要克服许多障碍,但科学家们已经迈出了关键的一步!在未来的研究中,加快神经恢复的方法学发展,以及建立新的用于控制神经系统特定细胞类型的工具,会让脊髓损伤的靶向和基于神经回路的治疗展现出更加光明的前景。 2.5 脊髓损伤后细胞簇的动态变化 脊髓损伤后的细胞相互作用及其细胞簇的动态变化对损伤脊髓的修复至关重要,这些发现可提高研究者们对脊髓损伤发展的理解,并为脊髓损伤在时间上治疗提供新的见解。细胞簇的动态变化特征见图6。"

| [1] FAN B, WEI Z, FENG S. Progression in translational research on spinal cord injury based on microenvironment imbalance. Bone Res. 2022; 10(1):35. [2] HUANG H, YOUNG W, SKAPER S, et al. Clinical neurorestorative therapeutic guidelines for spinal cord injury (IANR/CANR version 2019). J Orthop Translat. 2020;20:14-24. [3] JAMES SL, THEADOM A, ELLENBOGEN RG, et al. Global, regional, and national burden of traumatic brain injury and spinal cord injury, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(1):56-87. [4] JIANG B, SUN D, SUN H, et al. Prevalence, incidence, and external causes of traumatic spinal cord injury in China: a nationally representative cross-sectional survey. Front Neurol. 2021;12:784647. [5] BÁRBARA-BATALLER E, MÉNDEZ-SUÁREZ JL, ALEMÁN-SÁNCHEZ C, et al. Change in the profile of traumatic spinal cord injury over 15 years in Spain. Scand J Trauma Resusc Emerg Med. 2018;26(1):27. [6] O’SHEA TM, BURDA JE, SOFRONIEW MV. Cell biology of spinal cord injury and repair. J Clin Invest. 2017;127(9):3259-3270. [7] RIEW TR, JIN X, KIM S, et al. Temporal dynamics of cells expressing NG2 and platelet-derived growth factor receptor-β in the fibrotic scar formation after 3-nitropropionic acid-induced acute brain injury. Cell Tissue Res. 2021;385(3):539-555. [8] GUIMARÃES-CAMBOA N, CATTANEO P, SUN Y, et al. Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo. Cell Stem Cell. 2017;20(3):345-359.e5. [9] HILTON BJ, HUSCH A, SCHAFFRAN B, et al. An active vesicle priming machinery suppresses axon regeneration upon adult CNS injury. Neuron. 2022;110(1):51-69.e7. [10] EWAN EE, AVRAHAM O, CARLIN D, et al. Ascending dorsal column sensory neurons respond to spinal cord injury and downregulate genes related to lipid metabolism. Sci Rep. 2021;11(1):374. [11] SCHELKER M, FEAU S, DU J, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun. 2017; 8(1):2032. [12] CAO Y, ZHU S, YU B, et al. Single-cell RNA sequencing for traumatic spinal cord injury. FASEB J. 2022;36(12):e22656. [13] LI E, YAN R, YAN K, et al. Single-cell RNA sequencing reveals the role of immune-related autophagy in spinal cord injury in rats. Front Immunol. 2022;13:987344. [14] TANG F, BARBACIORU C, WANG Y, et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5): 377-382. [15] SASAGAWA Y, NIKAIDO I, HAYASHI T, et al. Quartz-seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14(4):R31. [16] HASHIMSHONY T, WAGNER F, SHER N, et al. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666-673. [17] JAITIN DA, KENIGSBERG E, KEREN-SHAUL H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776-779. [18] RAMSKÖLD D, LUO S, WANG YC, et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777-782. [19] PICELLI S, FARIDANI OR, BJÖRKLUND AK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171-181. [20] HAGEMANN-JENSEN M, ZIEGENHAIN C, CHEN P, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38(6):708-714. [21] MACOSKO EZ, BASU A, SATIJA R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202-1214. [22] KLEIN AM, MAZUTIS L, AKARTUNA I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015; 161(5):1187-1201. [23] JOVIC D, LIANG X, ZENG H, et al. Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med. 2022; 12(3):e694. [24] TANG X, HUANG Y, LEI J, et al. The single-cell sequencing: new developments and medical applications. Cell Biosci. 2019;9:53. [25] WANG S, SUN ST, ZHANG XY, et al. The evolution of single-cell RNA sequencing technology and application: progress and perspectives. Int J Mol Sci. 2023;24(3):2943. [26] SUN HJ, CHEN J, NI B, et al. Recent advances and current issues in single-cell sequencing of tumors. Cancer Lett. 2015;365(1):1-10. [27] MEREU E, LAFZI A, MOUTINHO C, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020; 38(6):747-755. [28] PARK ES, YAN JP, ANG RA, et al. Isolation and genome sequencing of individual circulating tumor cells using hydrogel encapsulation and laser capture microdissection. Lab Chip. 2018;18(12):1736-1749. [29] VAN DEN BRINK SC, SAGE F, VéRTESY Á, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14(10):935-936. [30] LAFZI A, MOUTINHO C, PICELLI S, et al. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat Protoc. 2018;13(12):2742-2757. [31] SHENG K, CAO W, NIU Y, et al. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat Methods. 2017;14(3): 267-270. [32] ZONG C, LU S, CHAPMAN AR, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622-1626. [33] DING J, ADICONIS X, SIMMONS SK, et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol. 2020;38(6):737-746. [34] ARGELAGUET R, ARNOL D, BREDIKHIN D, et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 2020;21(1):111. [35] SHALEK AK, SATIJA R, SHUGA J, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505): 363-369. [36] WAHANE S, ZHOU X, ZHOU X, et al. Diversified transcriptional responses of myeloid and glial cells in spinal cord injury shaped by HDAC3 activity. Sci Adv. 2021;7(9):eabd8811. [37] LI Y, HE X, KAWAGUCHI R, et al. Microglia-organized scar-free spinal cord repair in neonatal mice. Nature. 2020;587(7835):613-618. [38] WANG J, XU L, LIN W, et al. Single-cell transcriptome analysis reveals the immune heterogeneity and the repopulation of microglia by Hif1α in mice after spinal cord injury. Cell Death Dis. 2022;13(5):432. [39] HAKIM R, ZACHARIADIS V, SANKAVARAM SR, et al. Spinal cord injury induces permanent reprogramming of microglia into a disease-associated state which contributes to functional recovery. J Neurosci. 2021;41(40):8441-8459. [40] HOU J, BI H, GE Q, et al. Heterogeneity analysis of astrocytes following spinal cord injury at single-cell resolution. FASEB J. 2022;36(8):e22442. [41] FAN Y, WU X, HAN S, et al. Single-cell analysis reveals region-heterogeneous responses in rhesus monkey spinal cord with complete injury. Nat Commun. 2023;14(1):4796. [42] BATIUK MY, MARTIROSYAN A, WAHIS J, et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat Commun. 2020;11(1):1220. [43] WU Z, LI G, WANG S, et al. Single-cell analysis of spinal cord injury reveals functional heterogeneity of oligodendrocyte lineage cells. Gene. 2023;886:147713. [44] ZHANG Q, YU B, ZHANG Y, et al. Combination of single-cell and bulk RNA seq reveals the immune infiltration landscape and targeted therapeutic drugs in spinal cord injury. Front Immunol. 2023;14:1068359. [45] MILICH LM, CHOI JS, RYAN C, et al. Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord. J Exp Med. 2021;218(8):e20210040. [46] FISHER ES, AMARANTE MA, LOWRY N, et al. Single cell profiling of CD45+ spinal cord cells reveals microglial and B cell heterogeneity and crosstalk following spinal cord injury. J Neuroinflammation. 2022; 19(1):266. [47] SHU M, XUE X, NIE H, et al. Single-cell RNA sequencing reveals Nestin+ active neural stem cells outside the central canal after spinal cord injury. Sci China Life Sci. 2022;65(2):295-308. [48] STENUDD M, SABELSTRÖM H, LLORENS-BOBADILLA E, et al. Identification of a discrete subpopulation of spinal cord ependymal cells with neural stem cell properties. Cell Rep. 2022;38(9):110440. [49] SQUAIR JW, MILANO M, DE COUCY A, et al. Recovery of walking after paralysis by regenerating characterized neurons to their natural target region. Science. 2023;381(6664):1338-1345. [50] KATHE C, SKINNIDER MA, HUTSON TH, et al. The neurons that restore walking after paralysis. Nature. 2022;611(7936):540-547. [51] LIU A, YU L, LI X, et al. Celsr2-mediated morphological polarization and functional phenotype of reactive astrocytes in neural repair. Glia. 2023;71(8):1985-2004. [52] LIDDELOW SA, BARRES BA. Reactive astrocytes: production, function, and therapeutic potential. Immunity. 2017;46(6):957-967. [53] LIDDELOW SA, GUTTENPLAN KA, CLARKE LE, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017; 541(7638):481-487. [54] GAO MY, WANG JQ, HE J, et al. Single-cell RNA-sequencing in astrocyte development, heterogeneity, and disease. Cell Mol Neurobiol. 2023; 43(7):3449-3464. [55] ZHENG C, TU C, WANG J, et al. Deciphering oligodendrocyte lineages in the human fetal central nervous system using single-cell RNA sequencing. Mol Neurobiol. 2024;61(3):1737-1752. [56] SHANG J, MA C, DING H, et al. Development and validation of a differentiation-related signature based on single-cell RNA sequencing data of immune cells in spinal cord injury. Heliyon. 2023;9(9):e19853. [57] ANDERSON MA, O’SHEA TM, BURDA JE, et al. Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature. 2018;561(7723):396-400. |
| [1] | He Longcai, Song Wenxue, Ming Jiang, Chen Guangtang, Wang Junhao, Liao Yidong, Cui Junshuan, Xu Kaya. An experimental method for simultaneous extraction and culture of primary cortical neurons and microglial cells from SD rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1395-1400. |
| [2] | Zhao Nannan, Li Yanjie, Qin Hewei, Zhu Bochao, Ding Huimin, Xu Zhenhua. Changes in ferroptosis in hippocampal neurons of vascular dementia model rats treated with Tongmai Kaiqiao Pill [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1401-1407. |
| [3] | Chi Wenxin, Zhang Cunxin, Gao Kai, Lyu Chaoliang, Zhang Kefeng. Mechanism by which nobiletin inhibits inflammatory response of BV2 microglia [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1321-1327. |
| [4] | Zhang Shuai, Li Zichun, Xu Yihao, Xie Xiaofeng, Guo Zhongsheng, Zhao Qingyang. Effect of transcranial magneto-acousto-electrical stimulation on the plasticity of the prefrontal cortex network in mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1108-1117. |
| [5] | Zhao Ruihua, Chen Sixian, Guo Yang, Shi Lei, Wu Chengjie, Wu Mao, Yang Guanglu, Zhang Haoheng, Ma Yong. Wen-Shen-Tong-Du Decoction promoting spinal cord injury repair in mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1118-1126. |
| [6] | Liu Zhezhe, Yu Meiqing, Wang Tingting, Zhang Min, Li Baiyan. Troxerutin modulates nuclear factor-kappaB signaling pathway to inhibit brain injury and neuronal apoptosis in cerebral infarction rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1137-1143. |
| [7] | Wang Rongrong, Huang Yushan, Li Xiangmiao, Bai Jinzhu. Prostaglandin E1 regulates vascular-related factors and protects microcirculatory function during the acute phase of traumatic spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 958-967. |
| [8] | Lu Ranran, Zhou Xu, Zhang Lijie, Yang Xinling. Dimethyl fumarate alleviates nerve damage in a mouse model of Parkinson’s disease [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 989-994. |
| [9] | Yang Bin, Tao Guangyi, Yang Shun, Xu Junjie, Huang Junqing . Visualization analysis of research hotspots of artificial intelligence in field of spinal cord nerve injury and repair [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(4): 761-770. |
| [10] | Zheng Yitong, Wang Yongxin, Liu Wen, Amujite, Qin Hu. Action mechanism of intrathecal transplantation of human umbilical cord mesenchymal stem cell-derived exosomes for repair of spinal cord injury under neuroendoscopy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7743-7751. |
| [11] | Guo Jia, Ren Yafeng, Li Bing, Huang Jing, Shang Wenya, Yang Yike, Liu Huiyao. Action mechanism of mesenchymal stem cell-derived exosomes carrying miRNAs in improving spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7827-7838. |
| [12] | Xu Biao, Dong Yuzhen, Lu Tan. Effect of dihydroquercetin on the expression of inflammatory response markers in rats with spinal cord injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6843-6850. |
| [13] | Wang Ziheng, Wu Shuang. Oxidative stress-related genes and molecular mechanisms after spinal cord injury: data analysis and verification based on GEO database [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6893-6904. |
| [14] | Zhao Nan, Ding Yong, Xiu Hang, Liu Pengfei, Liang Guogang. Extraction and culture of enteric glial cells from C57BL/6 newborn neonatal mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6656-6660. |
| [15] | Wang Zhaoyan, Wang Qian, Liu Weipeng, Yang Hui, Luan Zuo, Qu Suqing. Effect of fibronectin on differentiation of human neural stem cells into oligodendrocyte precursor cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(31): 6661-6666. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||