Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (35): 7690-7700.doi: 10.12307/2025.973
Identification of immunodiagnostic biomarkers and drug screening for steroid-induced osteonecrosis of the femoral head
Han Jie1, Pan Chengzhen2, Shang Yuzhi1, Zhang Chi1
- 1Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China; 2Yulin Orthopedic Hospital of Integrated Traditional Chinese and Western Medicine, Yulin 537006, Guangxi Zhuang Autonomous Region, China
-
Received:2024-10-31Accepted:2024-12-31Online:2025-12-18Published:2025-05-07 -
Contact:Zhang Chi, MD, Attending physician, Department of Orthopedics, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China -
About author:Han Jie, Master, Professor, Chief physician, Department of Orthopedics, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, Nos. 82260858 and 82460872 (to HJ); Guangxi Natural Science Foundation, No. 2024GXNSFAA010243 (to HJ); Pilot Project for the High-Level Construction of Key Disciplines of Chinese Medicine in Guangxi, No. [2023]13 (to HJ); Guangxi Key Research Laboratory Construction Project of Traditional Chinese Medicine, No. [2023]9 (to HJ); Guangxi Young Qihuang Scholars Training Project, No. [2022]13 (to HJ); “Qihuang Engineering” High-Level Talent Team Cultivation Project of Guangxi University of Chinese Medicine, No. [2024]3 (to HJ)
CLC Number:
Cite this article
Han Jie, Pan Chengzhen, Shang Yuzhi, Zhang Chi. Identification of immunodiagnostic biomarkers and drug screening for steroid-induced osteonecrosis of the femoral head[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7690-7700.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
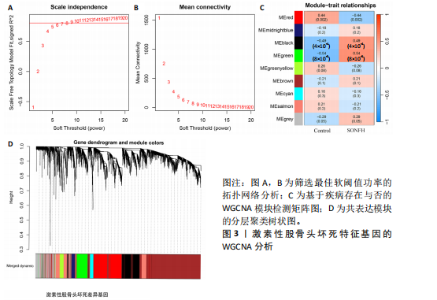
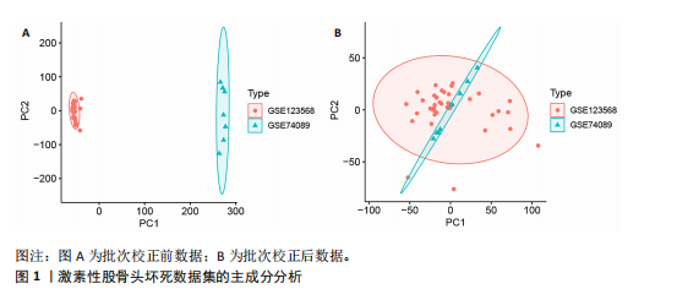
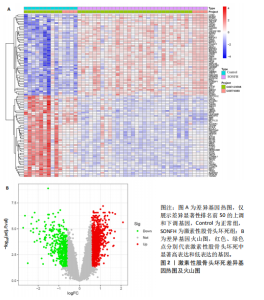
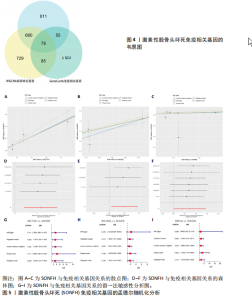
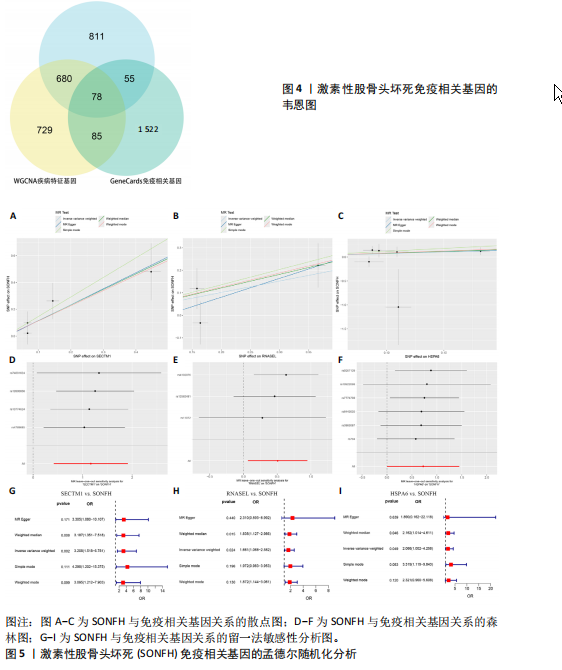
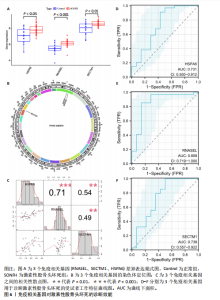
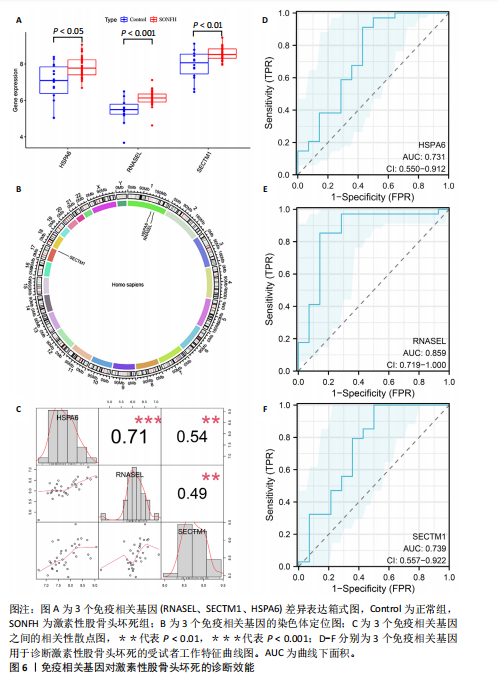
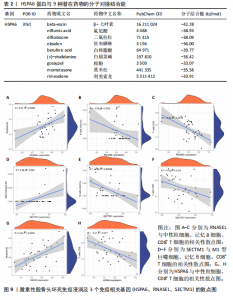
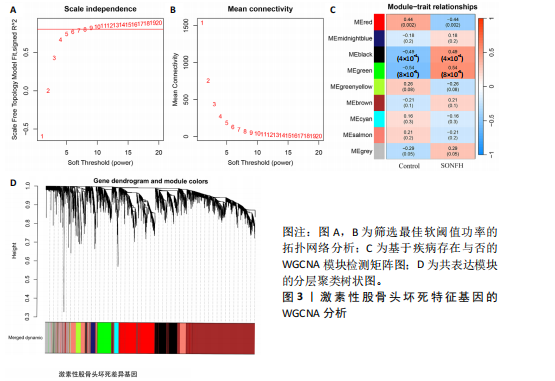
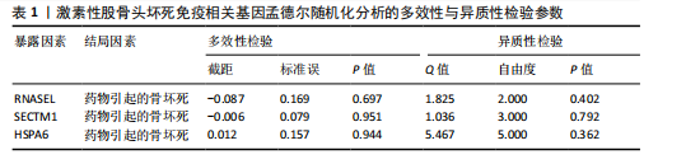
2.2 WGCNA分析激素性股骨头坏死的特征基因 采用WGCNA进一步校正后的全表达谱基因数据,该分析确定了9为最佳软阈值功率,确保基因网络中的无标度拓扑(图3A,B)。WGCNA结果显示绿色、黑色和红色模块的P均< 0.05,说明上述3个模块的基因与激素性股骨头坏死具有密切联系(图3C,D)。故将绿色、黑色和红色模块的306,506,760个基因取并集,得到1 572个激素性股骨头坏死的疾病特征基因,纳入后续研究。 2.3 激素性股骨头坏死免疫相关基因的筛选 从GeneCards数据库获得了1 740个免疫相关基因,将这些免疫相关基因与激素性股骨头坏死的1 624个差异基因和WGCNA的1 572个疾病特征基因取交集,共得到78个激素性股骨头坏死的免疫相关基因(图4)。 2.4 孟德尔随机化分析验证激素性股骨头坏死的免疫相关基因 孟德尔随机化分析结果表明,根据逆方差加权法从药物引起的骨坏死数据集中发现了3个与药物引起的骨坏死相关阳性基因(P < 0.05),分别为RNASEL(2-5A依赖性核糖核酸酶)、SECTM1(分泌型跨膜蛋白1)、HSPA6(热休克70 kDa蛋白6),它们均未出现显著异质性(P > 0.05),亦未出现显著多效性(P > 0.05),说明孟德尔随机化分析结果可靠(表1)。散点图表明RNASEL、SECTM1、HSPA6基因均是药物引起骨坏死的风险因素(图5A-C),留一法敏感性分析图显示去除SECTM1的任一单核苷酸多态性均不影响整体结果,而去除RNASEL或HSPA6的某些单核苷酸多态性可能会导致整体结果转变为阴性(图5D-F)。上述3个免疫相关基因与药物引起的骨坏死之间具有潜在因果关系(图5G-I)。 2.5 免疫相关基因的诊断效能与相关性分析 差异分析结果显示,与正常组相比,激素性股骨头坏死组中免疫相关基因RNASEL、SECTM1、HSPA6显著高表达(P < 0.05),见图6A。染色体定位图显示,HSPA6和RNASEL位于1号染色体上,SECTM1位于17和18号染色体之间,见图6B。相关性分析结果表明,激素性股骨头坏死样本中RNASEL、SECTM1、HSPA6基"
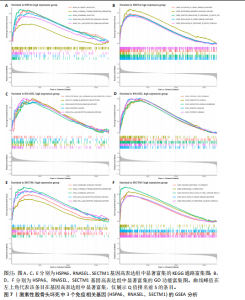
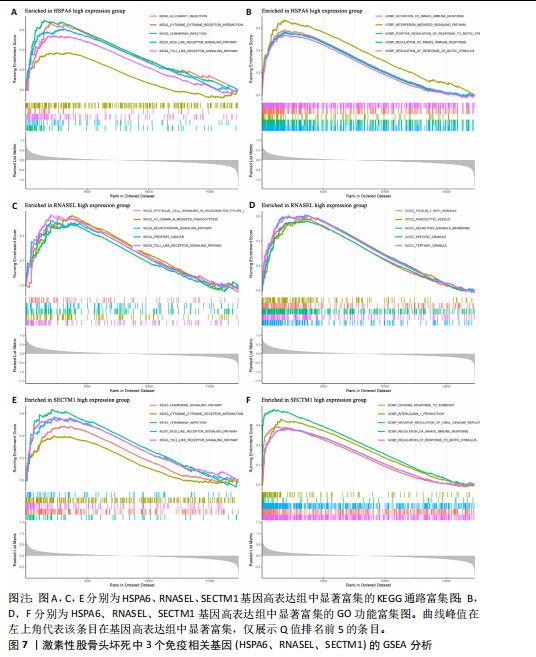
因的表达量两两之间均呈显著的正相关性(P < 0.01),见图6C。受试者工作特征曲线图显示上述3个基因在激素性股骨头坏死中的诊断效能均较高(曲线下面积AUC > 0.7),见图6D-F。 2.6 单个基因的GSEA分析 由于RNASEL、SECTM1、HSPA6在激素性股骨头坏死样本中显著高表达,因此进一步研究了RNASEL、SECTM1、HSPA6高表达组富集的相关通路与功能,进而分析上述3个基因在激素性股骨头坏死中的作用。GSEA富集结果表明,HSPA6高表达组涉及的KEGG信号通路包括NOD样受体信号通路、TOLL样受体信号通路、趋化因子信号通路、B细胞受体信号通路、 Janus激酶-信号转导与转录激活子信号通路、细胞周期、细胞凋亡等;生物学过程包括参与细胞对生物刺激的反应、白细胞介素1β生成的正向调节、白细胞介素8产生的正向调节、白细胞介素6的产生、淋巴细胞介导的免疫、炎症反应的正向调节等;细胞组分包括核糖体、典型炎性复合体、内吞囊泡等;分子功能包括TOLL样受体结合、未折叠蛋白结合、白细胞介素1受体结合、参与凋亡过程的肽酶激活剂活性等(图7A,B)。 RNASEL高表达组涉及的KEGG信号通路包括TOLL样受体信号通路、细胞黏附分子、胰岛素信号通路、细胞凋亡、NOD样受体信号通路、B细胞受体信号通路、T细胞受体信号通路、MTOR信号通路等;生物学过程包括干扰素介导的信号通路、白细胞介素1β生成的正向调节、对Ⅰ型干扰素的反应、干扰素β生成的正向调节、Ⅰ型干扰素产生的正向调节等;细胞组分包括核糖体、内吞囊泡、T细胞受体复合物等;分子功能包括核糖体的结构组成、TOLL样受体结合、白细胞介素1受体结合等(图7C,D)。 SECTM1高表达组涉及的KEGG信号通路包括TOLL样受体信号通路、NOD样受体信号通路、Janus激酶-信号转导与转录激活子信号通路、白细胞经内皮细胞迁移、细胞黏附分子、细胞周期、胰岛素信号通路等;生物学过程包括白细胞介素1β生成的正向调节、白细胞介素6产生的正向调节、白细胞介素8产生的正向调"
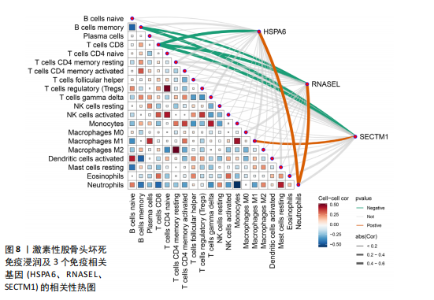
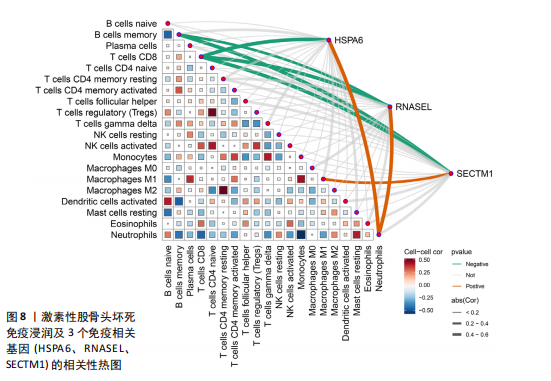
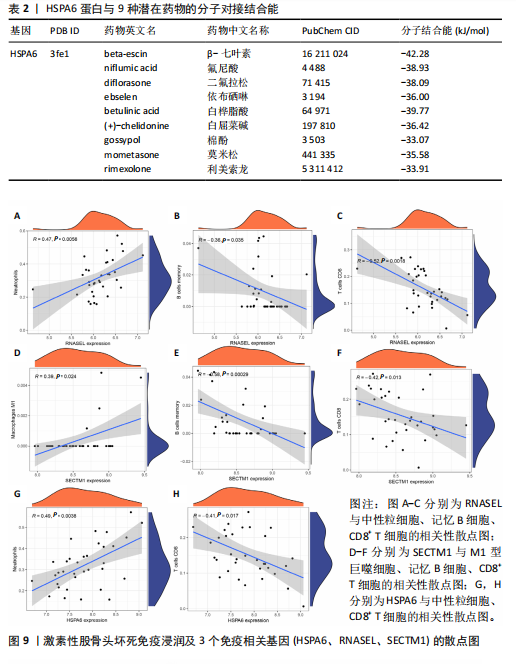
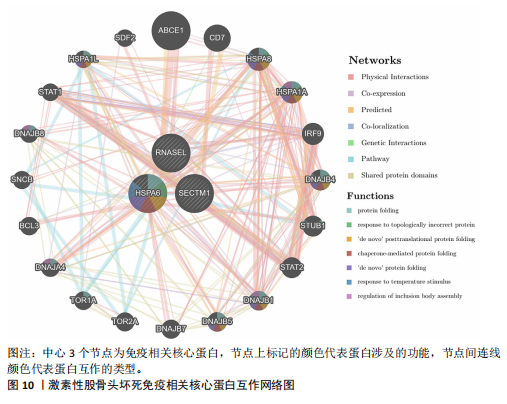
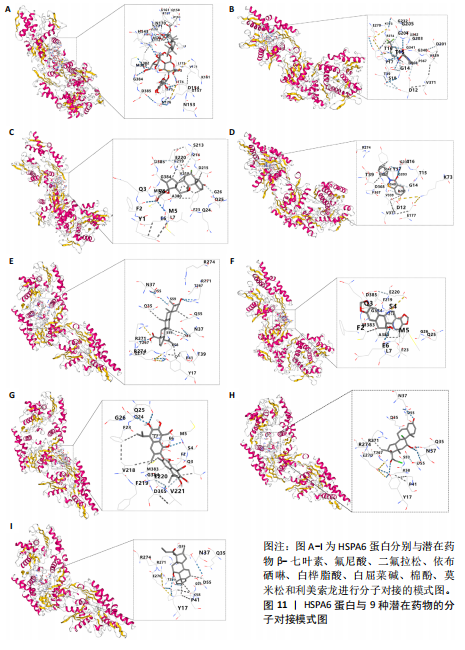
节、中性粒细胞趋化性、细胞对Ⅱ型干扰素的反应、Ⅰ型干扰素产生的正向调节等;细胞组分包括核糖体、内吞囊泡、典型炎性复合体等;分子功能包括TOLL样受体结合、白细胞介素1受体结合、未折叠蛋白结合等(图7E,F)。 2.7 免疫浸润相关性分析 结果显示RNASEL和HSPA6基因表达量均与中性粒细胞水平呈正相关(P < 0.05),RNASEL、SECTM1、HSPA6基因表达量与CD8+ T细胞水平呈负相关性(P < 0.05);RNASEL和SECTM1基因表达量与记忆B细胞水平呈负相关性(P < 0.05),SECTM1基因表达量与M1型巨噬细胞呈正相关(P < 0.05),见图8,9。 2.8 免疫相关蛋白互作网络 互作分析得到了由23个蛋白和305条互作关系组成的网络,其中包含77.64%物理互作关系、8.01%共表达互作关系、5.37%预测互作关系、3.63%共定位互作关系、2.87%遗传互作关系、1.88%通路互作关系、0.60%共享蛋白结构域互作关系。据统计有7个、5个、15个蛋白分别与RNASEL、SECTM1、HSPA6蛋白具有互作关系。其中,SECTM1与HSPA6之间具有共表达互作关系,与此次研究的免疫相关基因表达量相关性分析结果相一致。富集的相关功能包括蛋白质折叠、对Ⅰ型干扰素的细胞反应、泛素样蛋白连接酶结合、对细胞因子介导的信号通路的调节、对细胞因子刺激的反应调节等(图10),上述功能与此次研究GSEA富集的结果相类似。 2.9 潜在药物预测及分子对接 根据药物富集分析筛选标准,未富集到RNASEL和SECTM1基因的潜在药物;富集得到74种HSPA6基因的潜在药物(校正后P < 0.05)。为进一步验证潜在药物,将HSPA6蛋白与74种潜在药物进行分子对接,结果显示共有9种潜在药物和HSPA6蛋白的结合能< -29.3 kJ/mol,分别为β-七叶素、氟尼酸、二氟拉松、依布硒啉、白桦脂酸、白屈菜碱、棉酚、莫米松、利美索龙,其中β-七叶素的结合能最佳,为-42.28 kJ/mol(表2)。HSPA6蛋白与9种潜在药物的分子对接模型详见图11。"
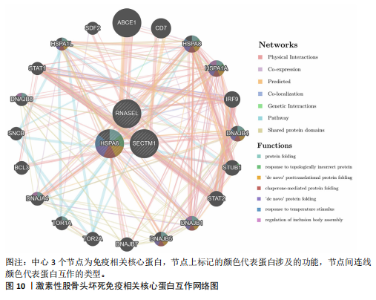
| [1] ZHAO DW, YU M, HU K, et al. Prevalence of Nontraumatic Osteonecrosis of the Femoral Head and its Associated Risk Factors in the Chinese Population: Results from a Nationally Representative Survey. Chin Med J (Engl). 2015;128(21):2843-2850. [2] 孙伟,高福强,李子荣.股骨头坏死临床诊疗技术专家共识(2022年)[J].中国修复重建外科杂志,2022,36(11):1319-1326. [3] ZHAO D, ZHANG F, WANG B, et al. Guidelines for clinical diagnosis and treatment of osteonecrosis of the femoral head in adults (2019 version). J Orthop Translat. 2020; 21:100-110. [4] MUSTAFA SS. Steroid-induced secondary immune deficiency. Ann Allergy Asthma Immunol. 2023;130(6):713-717. [5] POWELL C, CHANG C, GERSHWIN ME. Current concepts on the pathogenesis and natural history of steroid-induced osteonecrosis. Clin Rev Allergy Immunol. 2011;41(1):102-113. [6] KWON H M, HAN M, LEE TS, et al. Effect of Corticosteroid Use on the Occurrence and Progression of Osteonecrosis of the Femoral Head: A Nationwide Nested Case-Control Study. J Arthroplasty. 2024;39(10):2496-2505. [7] WU T, SHI W, ZHOU Y, et al. Identification and validation of endoplasmic reticulum stress-related genes in patients with steroid-induced osteonecrosis of the femoral head. Sci Rep. 2024;14(1):21634. [8] YANG S, ZHAO Y, TAN Y, et al. Identification of microtubule-associated biomarker using machine learning methods in osteonecrosis of the femoral head and osteosarcoma. Heliyon. 2024;10(11):e31853. [9] ZHU Y, WANG X, LIU R. Bioinformatics proved the existence of potential hub genes activating autophagy to participate in cartilage degeneration in osteonecrosis of the femoral head. J Mol Histol. 2024; 55(4):539-554. [10] AN W, YANG Y, HE W, et al. Three-dimensional mapping of necrotic lesions for early-stage osteonecrosis of the femoral head. J Orthop Surg Res. 2024;19(1):577. [11] 李岩,王兴河.基于生物信息学分析三阴性乳腺癌的差异表达基因和药物靶点挖掘[J].中国临床药理学杂志,2024,40(2): 269-272. [12] SEKULA P, DEL GMF, PATTARO C, et al. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc Nephrol. 2016;27(11):3253-3265. [13] 林苗远,杨继滨,闫文强,等.组织工程技术促进股骨头坏死骨组织再血管化的研究进展[J].中国修复重建外科杂志, 2021,35(11):1479-1485. [14] JIA Y, ZHANG Y, LI S, et al. Identification and assessment of novel dynamic biomarkers for monitoring non-traumatic osteonecrosis of the femoral head staging. Clin Transl Med. 2023;13(6): 1295. [15] LU X, WANG X, WANG P, et al. Identification of candidate genes and chemicals associated with osteonecrosis of femoral head by multiomics studies and chemical-gene interaction analysis. Front Endocrinol (Lausanne). 2024;15:1419742. [16] GONG N, TUO Y, LIU P. Identification and Mendelian randomization validation of pathogenic gene biomarkers in obstructive sleep apnea. Front Neurol. 2024;15:1442835. [17] GHARIB SA, HURLEY AL, ROSEN MJ, et al. Obstructive sleep apnea and CPAP therapy alter distinct transcriptional programs in subcutaneous fat tissue. Sleep. 2020;43(6): zsz314. [18] LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [19] WANG Q, SU Z, ZHANG J, et al. Unraveling the copper-death connection: Decoding COVID-19’s immune landscape through advanced bioinformatics and machine learning approaches. Hum Vaccin Immunother. 2024;20(1):2310359. [20] JI L, WANG Y, LU T, et al. Assessing the causal relationship between blood metabolites and low back pain: a Mendelian randomization study. Am J Transl Res. 2024;16(4):1366-1374. [21] KURKI MI, KARJALAINEN J, PALTA P, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508-518. [22] BURGESS S, DUDBRIDGE F, THOMPSON SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880-1906. [23] HEMANI G, BOWDEN J, DAVEY SG. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27(R2):R195-R208. [24] SUN Q, BAI L, ZHU S, et al. Analysis of Lymphoma-Related Genes with Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment. Biomed Res Int. 2022;2022:8503511. [25] LV Y, JI L, DAI H, et al. Identification of key regulatory genes involved in myelination after spinal cord injury by GSEA analysis. Exp Neurol. 2024;382:114966. [26] GENTLES AJ, NEWMAN AM, LIU CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938-945. [27] WARDE-FARLEY D, DONALDSON SL, COMES O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214-W220. [28] YOO M, SHIN J, KIM J, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31(18):3069-3071. [29] TROTT O, OLSON AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455-461. [30] 李洵珣,金晨,陈康,等.基于网络药理学和分子对接分析丰城鸡血藤治疗乳腺癌的分子靶点和机制[J]. 中国药理学通报,2022,38(5):767-775. [31] LI XH, PANG WW, ZHANG Y, et al. A Mendelian randomization study for drug repurposing reveals bezafibrate and fenofibric acid as potential osteoporosis treatments. Front Pharmacol. 2023;14: 1211302. [32] HUANG X, ZHANG T, GUO P, et al. Association of antihypertensive drugs with fracture and bone mineral density: A comprehensive drug-target Mendelian randomization study. Front Endocrinol (Lausanne). 2023;14:1164387. [33] GU Y, JIN Q, HU J, et al. Causality of genetically determined metabolites and metabolic pathways on osteoarthritis: a two-sample mendelian randomization study. J Transl Med. 2023;21(1): 357. [34] BORGELT L, HAACKE N, LAMPE P, et al. Small-molecule screening of ribonuclease L binders for RNA degradation. Biomed Pharmacother. 2022;154:113589. [35] FERRANDI C, RICHARD F, TAVANO P, et al. Characterization of immune cell subsets during the active phase of multiple sclerosis reveals disease and c-Jun N-terminal kinase pathway biomarkers. Mult Scler. 2011;17(1):43-56. [36] WANG T, HUANG C, LOPEZ-CORAL A, et al. K12/SECTM1, an interferon-γ regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J Leukoc Biol. 2012;91(3):449-459. [37] WANG T, GE Y, XIAO M, et al. SECTM1 produced by tumor cells attracts human monocytes via CD7-mediated activation of the PI3K pathway. J Invest Dermatol. 2014;134(4):1108-1118. [38] SONG B, SHEN S, FU S, et al. HSPA6 and its role in cancers and other diseases. Mol Biol Rep. 2022;49(11):10565-10577. [39] JIANG J, LIU X, LAI B, et al. Correlational analysis between neutrophil granulocyte levels and osteonecrosis of the femoral head. BMC Musculoskelet Disord. 2019;20(1):393. [40] NONOKAWA M, SHIMIZU T, YOSHINARI M, et al. Association of Neutrophil Extracellular Traps with the Development of Idiopathic Osteonecrosis of the Femoral Head. Am J Pathol. 2020;190(11):2282-2289. [41] LUO D, GAO X, ZHU X, et al. Identification of steroid-induced osteonecrosis of the femoral head biomarkers based on immunization and animal experiments. BMC Musculoskelet Disord. 2024;25(1):596. [42] CHEN B, LIU Y, CHENG L. IL-21 Enhances the Degradation of Cartilage Through the JAK-STAT Signaling Pathway During Osteonecrosis of Femoral Head Cartilage. Inflammation. 2018;41(2):595-605. [43] GENG W, ZHANG W, MA J. IL-9 exhibits elevated expression in osteonecrosis of femoral head patients and promotes cartilage degradation through activation of JAK-STAT signaling in vitro. Int Immunopharmacol. 2018;60:228-234. [44] KOSUKEGAWA I, OKAZAKI S, YAMAMOTO M, et al. The proton pump inhibitor, lansoprazole, prevents the development of non-traumatic osteonecrosis of the femoral head: an experimental and prospective clinical trial. Eur J Orthop Surg Traumatol. 2020;30(4):713-721. [45] ADAPALA NS, YAMAGUCHI R, PHIPPS M, et al. Necrotic Bone Stimulates Proinflammatory Responses in Macrophages through the Activation of Toll-Like Receptor 4. Am J Pathol. 2016;186(11): 2987-2999. [46] YU R, MA C, LI G, et al. Inhibition of Toll-Like Receptor 4 Signaling Pathway Accelerates the Repair of Avascular Necrosis of Femoral Epiphysis through Regulating Macrophage Polarization in Perthes Disease. Tissue Eng Regen Med. 2023;20(3):489-501. [47] HE Z, HU Y, ZHANG Y, et al. Asiaticoside exerts neuroprotection through targeting NLRP3 inflammasome activation. Phytomedicine. 2024;127:155494. [48] BARROW F, KHAN S, FREDRICKSON G, et al. Microbiota-Driven Activation of Intrahepatic B Cells Aggravates NASH Through Innate and Adaptive Signaling. Hepatology. 2021; 74(2):704-722. [49] CEGIEŁA U, PYTLIK M, JANIEC W, et al. Effects of alpha-Escin on mechanical features of the femoral bone in rats with experimental post-steroid osteopenia. Acta Pol Pharm. 2000;57(4):317-320. |
| [1] | Zhang Yibo, Lu Jianqi, Mao Meiling, Pang Yan, Dong Li, Yang Shangbing, Xiao Xiang. Exploring the causal relationship between rheumatoid arthritis and coronary atherosclerosis: a Mendel randomized study involving serum metabolites and inflammatory factors [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-9. |
| [2] | Chen Jiayong, Tang Meiling, Lu Jianqi, Pang Yan, Yang Shangbing, Mao Meiling, Luo Wenkuan, Lu Wei, Zhou Jiatan. Based on Mendelian randomization, the causal relationship between 1400 metabolites and sarcopenia and the correlation analysis of cardiovascular disease were investigated [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-11. |
| [3] | Dong Tingting, Chen Tianxin, Li Yan, Zhang Sheng, Zhang Lei. Causal relationship between modifiable factors and joint sports injuries [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1953-1962. |
| [4] | Chen Shuai, Jin Jie, Han Huawei, Tian Ningsheng, Li Zhiwei . Causal relationship between circulating inflammatory cytokines and bone mineral density based on two-sample Mendelian randomization [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1556-1564. |
| [5] |
Zhao Wensheng, Li Xiaolin, Peng Changhua, Deng Jia, Sheng Hao, Chen Hongwei, Zhang Chaoju, He Chuan.
Gut microbiota and osteoporotic fractures #br#
#br#
[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1296-1304.
|
| [6] | Ma Haoyu, Qiao Hongchao, Hao Qianqian, Shi Dongbo. Causal effects of different exercise intensities on the risk of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1305-1311. |
| [7] | Li Jiatong, Jin Yue, Liu Runjia, Song Bowen, Zhu Xiaoqian, Li Nianhu . Association between thyroid function levels and phenotypes associated with sarcopenia [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1312-1320. |
| [8] | Wu Guangtao, Qin Gang, He Kaiyi, Fan Yidong, Li Weicai, Zhu Baogang, Cao Ying . Causal relationship between immune cells and knee osteoarthritis: a two-sample bi-directional Mendelian randomization analysis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1081-1090. |
| [9] | Fang Yuan, Qian Zhiyong, He Yuanhada, Wang Haiyan, Sha Lirong, Li Xiaohe, Liu Jing, He Yachao, Zhang Kai, Temribagen. Mechanism of Mongolian medicine Echinops sphaerocephalus L. in proliferation and angiogenesis of vascular endothelial cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7519-7528. |
| [10] | Li Chen, Liu Ye, Ni Xindi, Zhang Yuang. Simulation analysis of real-time continuous stiffness in muscle fibers and tendons of the triceps surae during multi-joint movement [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7529-7536. |
| [11] | Yang Bo, Pan Xinfang, Chang Liuhui, Ni Yong. Correlation of echocardiographic parameters with disability at 3 months after acute ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7544-7551. |
| [12] | Liu Xuan, Ding Yuqing, Xia Ruohan, Wang Xianwang, Hu Shujuan. Exercise prevention and treatment of insulin resistance: role and molecular mechanism of Keap1/nuclear factor erythroid2-related factor 2 signaling pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7578-7588. |
| [13] | Wang Xuepeng, , He Yong, . Effect of insulin-like growth factor family member levels on inflammatory arthritis: a FinnGen biobank-based analysis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7656-7662. |
| [14] | Wang Tao, Wang Shunpu, Min Youjiang, Wang Min, Li Le, Zhang Chen, Xiao Weiping. Causal relationship between gut microbiota and rheumatoid arthritis: data analysis in European populations based on GWAS data [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7663-7668. |
| [15] | Gong Yuehong, Wang Mengjun, Ren Hang, Zheng Hui, Sun Jiajia, Liu Junpeng, Zhang Fei, Yang Jianhua, Hu Junping. Machine learning combined with bioinformatics screening of key genes for pulmonary fibrosis associated with cellular autophagy and experimental validation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7679-7689. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||