Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (32): 7028-7040.doi: 10.12307/2025.937
Screening of target genes for bile acid metabolism in Crohn’s disease and its value in disease diagnosis and therapeutic monitoring
Chen Hui1, Zhang Lu1, Chen Jing1, Wang Nanzhang1, Wang Ruochun1, Lu Cuihua1, 2, Ji Yifei1, 2
- 1Department of Gastroenterology, 2Clinical Medical Research Center, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China
-
Received:2024-10-16Accepted:2024-12-10Online:2025-11-18Published:2025-04-29 -
Contact:Ji Yifei, Associate chief physician, Master’s supervisor, Department of Gastroenterology, and Clinical Medical Research Center, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China -
About author:Chen Hui, Master candidate, Department of Gastroenterology, Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China -
Supported by:National Natural Science Foundation of China, No. 82070624 (to LCH); Jiangsu Provincial Health Commission Research Project, No. H2019072 (to JYF); Jiangsu Provincial Science and Technology Project, No. BE2019692 (to JYF); Nantong Municipal Health Commission Research Project, No. MSZ2023004 (to JYF)
CLC Number:
Cite this article
Chen Hui, Zhang Lu, Chen Jing, Wang Nanzhang, Wang Ruochun, Lu Cuihua, Ji Yifei. Screening of target genes for bile acid metabolism in Crohn’s disease and its value in disease diagnosis and therapeutic monitoring[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 7028-7040.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

2.1 生物制剂治疗克罗恩病后胆汁酸代谢相关基因表达恢复 对第1组差异表达的604个基因(图1A)与第2组差异表达的593个基因(图1B)取交集,获得324个在两组中变化方向相同的交叉基因,包括152个上调基因、172个下调基因。324个交叉基因在炎症组、治疗组和对照组中的差异表达显示在热图(图1C)中。KEGG富集分析结果显示,在11个胆汁酸代谢相关交集差异基因中,CYP3A4、SULT2A1、UGT2A3在胆汁酸代谢通路中的作用被标记为胆汁酸结合与解毒,ABCC2、SLC51B、SLC51A被标记为胆汁酸转运,ABCB1、ABCG2、SLC5A1涉及阳离子、类固醇、葡萄糖的转运,这9个基因表达在生物制剂治疗后恢复。此外,克罗恩病肠道炎症状态下CA2、AQP9表达上调,并在治疗后回落至正常状态。 2.2 GO、KEGG、GSEA分析揭示生物制剂治疗对克罗恩病生物学过程和胆汁酸相关代谢通路的影响 对324个交叉差异基因进行功能富集分析,结"


果见图2。GO分析显示,152个上调基因在以下生物学过程中富集:粒细胞趋化性、粒细胞迁移、髓系白细胞迁移、白细胞趋化性、中性粒细胞迁移、中性粒细胞趋化性;KEGG分析显示,152个上调基因在以下代谢通路中富集:肌肉细胞中的细胞骨架、细胞因子-细胞因子受体相互作用、 磷脂酰肌醇3-激酶-蛋白激酶B信号通路、阿米巴病、局部黏附。GO分析显示,172个下调基因在以下生物学过程中富集:有机阴离子运输、羧酸运输、异物代谢过程、有机酸运输、肠道吸收;KEGG分析显示,172个下调基因在以下代谢通路中富集:蛋白质消化和吸收、脂肪消化和吸收、胆汁分泌、视黄醇代谢、化学致癌-DNA加合物等。总的来说,经生物制剂治疗后,克罗恩病患者的肠道营养物质吸收以及胆汁酸代谢等多种物质代谢过程有一定程度恢复,白细胞趋化运动及炎症反应途径经治疗后的回落也在意料之中。对第1组和第2组差异表达基因分别进行GSEA富集分析,发现5条在对照组及治疗组显著上调的胆汁酸代谢相关信号通路(图2C,D),包括药物诱导胆汁酸通路、胆汁酸和胆盐的循环利用、胆汁循环、胆汁酸合成和胆汁运输障碍、葡萄糖醛化。关键基因UGT2A3位于葡萄糖醛化(REACTOME_GLUCURONIDATION)途径中,这进一步验证了生物制剂治疗有益于胆汁酸代谢相关的多个通路的恢复。 2.3 筛选胆汁酸代谢相关核心差异基因 基于STRING 12.0中的蛋白质互作数据,在Cytoscape中绘制蛋白质互作网络(图3),在Centiscape2.2插件中,利用中介中心性、接近中心性、度中心性3种算法筛选出324个差异基因中评分最高的51个关键基因(表1),其中包含胆汁酸代谢相关交集差异基因的大部分基因:ABCC2、CYP3A4、UGT2A3、ABCB1、ABCG2、SLC5A1,这从侧面证明了胆汁酸代谢相关交集差异基因 的关键性。 2.4 UGT2A3为生物制剂治疗响应密切的胆汁酸代谢相关核心基因 利用WCGNA识别共表达模块,当软阈值设置为8时,邻接函数能够较好地满足无尺度条件(图4A),得到了19个与治疗前后状态相关的模块(图4B),计算每个颜色模块的特征值与临床状态(未经生物制剂治疗与生物制剂治疗后)之间的相关系数和相关性显著性,筛选出了与生物制剂治疗响应密切的蓝色模块(-0.54,P < 0.000 1)、粉色模块(-0.58,P < 0.000 1),见图4C,共包含481个基因。在胆汁酸代谢相关交集差异基因中,只有UGT2A3对生物制剂治疗显示出了较强的响应(图4D),并且UGT2A3也作为蛋白质互作网络的关键基因,其在治疗后的表达恢复可能与生物制剂作用机制相偶联。 2.5 UGT2A3在炎症肠道组织中明显下调且经生物制剂治疗后表达恢复 对GSE134809数据集进行单细胞分析细胞聚类,使用JackStrawPlot功能可视化确定主成分个数的结果,确定显著性最大的主成分数为20(图5A,B),观察了UGT2A3在不同样本和细胞群间的表达情况,发现其表达主要集中在肠上皮细胞、杯状细胞,并且炎症组与对照组相比UGT2A3的表达密度明显减少(图5C-F),这些结果在临床炎症组结肠炎症标本的免疫组化实验中得到验证(图5G-H)。在GSE186582数据集炎症组、对照组、治疗组的UGT2A3表达箱式图中,炎症组UGT2A3表达明显下调,治疗组UGT2A3表达恢复(图6A)。通过临床结肠组织样本进一步验证了UGT2A3在炎症组、治疗组、对照组中的表达,Western blot检测(图6B,C)和qRT-PCR检测(图6D)显示炎症组织中UGT2A3显著低表达,并且经生物制剂治疗后UGT2A3表达恢复。 2.6 UGT2A3表达与克罗恩病患者临床数据的相关性及诊断能力 相关性"


分析表明,UGT2A3与全身炎症反应和克罗恩病的活动性密切相关(图7A-D),UGT2A3与C-反应蛋白呈强负相关(r=-0.798 0,P < 0.000 1),UGT2A3与血细胞沉降率呈中等程度负相关(r=-0.647 9,P < 0.05),UGT2A3与克罗恩病活动指数呈中等程度负相关(r=-0.584 5,P < 0.000 1),UGT2A3与克罗恩病内镜活动评分呈中等程度负相关(r=-0.584 7,P < 0.000 1)。针对UGT2A3高、低表达组临床数据对比结果显示,两组在性别、年龄、体质量指数、病程、吸烟史、伴随疾病、肛瘘史方面比较无明显差异(P > 0.05),见表2。 与UGT2A3高表达组相比,UGT2A3低表达组临床患者显示出了更多的粪隐血可能性、更严重的穿透性炎症、肠道狭窄等,见表3。粪便代谢组学分析发现,对照组初级胆汁酸水平明显低于炎症组(P < 0.000 1),次级胆汁酸水平明显高于炎症组(P < 0.000 1), 见图7E。相关性分析显示,UGT2A3的表达与初级胆汁酸水平呈强正相关(r=0.757 2,P < 0.05),与次级胆汁酸水平呈强负相关(r=-0.745 8,P < 0.05),见图7F-G。炎症组与对照组、炎症组与治疗组的受试者工作特征曲线显示UGT2A3具有良好的诊断能力(曲线下面积AUC=0.801 0),见图7H,并且能有效反映治疗带来的变化(曲线下面积AUC=0.800 8),见图7I。 2.7 胆汁酸相关基因与免疫浸润的相关性及UGT2A3高低表达组免疫浸润分析 对GSE186582数据集的炎症组样本进行免疫细胞浸润分析,得到上调的9个胆汁酸代谢相关交集差异基因与免疫细胞之间的相关性热图(图8A),结果提示黑素细胞、内皮细胞、微血管内皮细胞、成纤维细胞、单核细胞、周细胞、淋巴管内皮细胞、前脂肪细胞与UGT2A3负相关性较强(P < 0.000 1)。UGT2A3与免疫评分的相关性分析显示,UGT2A3表达与免疫评分(ImmuneScore)中度相关(r=-0.515,P < 0.000 1),与微环境评分(MicroenvironmentScore)高度相关(r=-0.718,P < 0.000 1),与基质评分(StromaScore)中度相关(r=-0.649,P < 0.000 1),见图8B-D。将炎症组分"


为UGT2A3高、低表达组,分析各组免疫细胞的表达水平(图9A-C),结果显示UGT2A3低表达组中抗原提呈树突状细胞(aDC)、B细胞(B cells)、CD8+原始T细胞(CD8+ naive T-cells)、类别转换记忆B细胞(Class-switched memory B-cells)、内皮细胞(Endothelial cells)、成纤维细胞(Fibroblasts)、淋巴管内皮细胞(ly Endothelial cells)、记忆B细胞(Memory B-cells)、单核细胞(Monocytes)、微血管内皮细胞(mv Endothelial cells)、原始B细胞(naive B-cells)、中性粒细胞(Neutrophils)、周细胞(Pericytes)、B祖细胞(pro B-cells)、Th1细胞(Th1 cells)更丰富(P < 0.000 1)。综上所述,UGT2A3低表达组具有更高的免疫浸润水平,UGT2A3与内皮细胞、周细胞、成纤维细胞和前脂肪细胞的密切相关性意味着其可能参与克罗恩病纤维化过程或受到维化的影响,具有进一步研究的价值。"


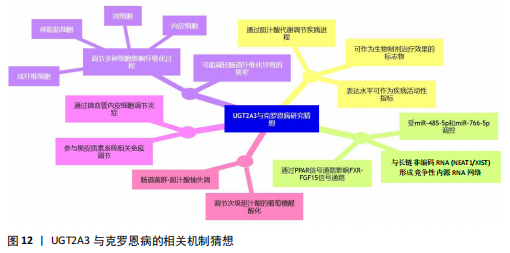
2.8 UGT2A3的mRNA-miRNA-lncRNA网络构建及调控miRNA功能富集结果 为了对UGT2A3参与生物代谢途径的机制进行研究,对UGT2A3调控的miRNA进行了功能富集分析(图10),结果显示结缔组织生长因子产生、成纤维细胞生长因子产生、生长激素分泌细胞分化等生物过程上调,这一定程度上解释了UGT2A3参与肠道纤维化的可能机制。另外,蛋白酶激活受体信号通路、调控干细胞多能性的信号通路、缺氧诱导因子1信号类风湿关节炎通路的显著富集也为UGT2A3和克罗恩病炎症的更深层次的研究提供了方向。通过构建UGT2A3的上游调控miRNA和lncRNA网络(图11)发现,hsa-miR-485-5p、hsa-miR-766-5p与UGT2A3之间具有最强的相关性,2个lncRNA显示出了与更多的miRNA节点相连接,这代表着hsa-miR-485-5p、hsa-miR-766-5p、NEAT1、XIST在该网络中的核心地位。"

| [1] RODA G, CHIEN NG S, KOTZE PG, et al. Crohn’s disease. Nat Rev Dis Primers. 2020;6(1):22. [2] LAMB CA, KENNEDY NA, RAINE T, et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults . Gut. 2019;68(Suppl 3):s1-s106. [3] HANAUER SB, FEAGAN BG, LICHTENSTEIN GR, et al. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet. 2002;359(9317):1541-1519. [4] SCHNITZLER F, FIDDER H, FERRANTE M, et al. Long-term outcome of treatment with infliximab in 614 patients with Crohn’s disease: results from a single-centre cohort. Gut. 2009;58(4):492-500. [5] FERNANDES P, SHARMA Y, ZULQARNAIN F, et al. Identifying metabolic shifts in Crohn’s disease using’ omics-driven contextualized computational metabolic network models. Sci Rep. 2023;13(1): 203. [6] DI CIAULA A, BONFRATE L, KHALIL M, et al. The interaction of bile acids and gut inflammation influences the pathogenesis of inflammatory bowel disease. Intern Emerg Med. 2023;18(8): 2181-2197. [7] LLOYD-PRICE J, ARZE C, ANANTHAKRISHNAN AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019; 569(7758):655-662. [8] VICH VILA A, ZHANG J, LIU M, et al. Untargeted faecal metabolomics for the discovery of biomarkers and treatment targets for inflammatory bowel diseases. Gut. 2024;73(11): 1909-1920. [9] LAVELLE A, SOKOL H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223-237. [10] BIAGIOLI M, MARCHIANO S, CARINO A, et al. Bile Acids Activated Receptors in Inflammatory Bowel Disease. Cells. 2021;10(6):1281. [11] JIN W, ZHENG M, CHEN Y, et al. Update on the development of TGR5 agonists for human diseases. Eur J Med Chem. 2024;271:116462. [12] SHIN DJ, WANG L. Bile Acid-Activated Receptors: A Review on FXR and Other Nuclear Receptors. Handb Exp Pharmacol. 2019;256:51-72. [13] MA Y, YANG H, WANG X, et al. Bile acids as signaling molecules in inflammatory bowel disease: Implications for treatment strategies. J Ethnopharmacol. 2025;337(Pt 3):118968. [14] NGOLLO M, PEREZ K, HAMMOUDI N, et al. Identification of Gene Expression Profiles Associated with an Increased Risk of Post-Operative Recurrence in Crohn’s Disease. J Crohns Colitis. 2022;16(8):1269-1280. [15] RITCHIE ME, PHIPSON B, WU D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [16] BARDOU P, MARIETTE J, ESCUDIÉ F, et al. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15(1):293. [17] YU G, WANG L G, HAN Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284-287. [18] LUO W, BROUWER C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics (Oxford, England). 2013;29(14):1830-1831. [19] LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. [20] LANGFELDER P, HORVATH S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46(11):i11. [21] SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. [22] BUTLER A, HOFFMAN P, SMIBERT P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411-420. [23] LALL S, SINHA D, BANDYOPADHYAY S, et al. Structure-Aware Principal Component Analysis for Single-Cell RNA-seq Data. J Comput Biol. 2018; 25(12):1365-1373. [24] PONT F, TOSOLINI M, FOURNIÉ JJ. Single-Cell Signature Explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res. 2019;47(21):e133. [25] QUAN F, LIANG X, CHENG M, et al. Annotation of cell types (ACT): a convenient web server for cell type annotation. Genome Med. 2023;15(1):91. [26] SUN J, DING J, YUE H, et al. Hypoxia-induced BNIP3 facilitates the progression and metastasis of uveal melanoma by driving metabolic reprogramming. Autophagy. 2024:1-19.doi: 10.1080/15548627.2024.2395142. [27] ARAN D, HU Z, BUTTE AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [28] TSUJI Y, HUANG Y, PENG J, et al. Identification of key ferroptosis genes in diabetic retinopathy based on bioinformatics analysis. PLoS One. 2023; 18(1):e0280548. [29] APARICIO-PUERTA E, HIRSCH P, SCHMARTZ GP, et al. miEAA 2023: updates, new functional microRNA sets and improved enrichment visualizations. Nucleic Acids Res. 2023;51(W1):W319-W325. [30] DOSEDELOVA V, ITTERHEIMOVA P, KUBAN P. Analysis of bile acids in human biological samples by microcolumn separation techniques: A review. Electrophoresis. 2021;42(1-2):68-85. [31] 胡静怡,沈洪,朱磊,等.胆汁酸在炎症性肠病中作用的研究进展[J].东南大学学报(医学版), 2021,40(1):108-113. [32] DING NS, MCDONALD JAK, PERDONES-MONTERO A, et al. Metabonomics and the Gut Microbiome Associated With Primary Response to Anti-TNF Therapy in Crohn’s Disease. J Crohns Colitis. 2020; 14(8):1090-1102. [33] GOTOH-SAITO S, ABE T, FURUKAWA Y, et al. Characterization of human UGT2A3 expression using a prepared specific antibody against UGT2A3. Drug Metab Pharmacokinet. 2019;34(4): 280-286. [34] HU DG, MARRI S, MACKENZIE PI, et al. The Expression Profiles and Deregulation of UDP-Glycosyltransferase (UGT) Genes in Human Cancers and Their Association with Clinical Outcomes. Cancers (Basel). 2021;13(17):4491. [35] YANG X, LI P, ZHUANG J, et al. Identification of Molecular Targets of Bile Acids Acting on Colorectal Cancer and Their Correlation with Immunity. Dig Dis Sci. 2024;69(1):123-134. [36] 李宏峰,赵俊芳,陈雪雯,等.UGT2A3差异表达在结直肠癌发生和早期诊断中的作用机制[J].国际生物医学工程杂志,2021,44(3):184-191. [37] 高源.UGT2A3调控结肠癌增殖及迁移的作用及其机制研究[D].广州:南方医科大学,2020. [38] PANG B, XU X, LU Y, et al. Prediction of new targets and mechanisms for quercetin in the treatment of pancreatic cancer, colon cancer, and rectal cancer . Food Funct. 2019;10(9): 5339-5349. [39] SALIMY S, LANJANIAN H, ABBASI K, et al. A deep learning-based framework for predicting survival-associated groups in colon cancer by integrating multi-omics and clinical data. Heliyon. 2023;9(7):e17653. [40] ZHAO Q, WANG N, LI Y, et al. [Lnc-TMEM132D-AS1 overexpression reduces sensitivity of non-small cell lung cancer cells to osimertinib]. Nan Fang Yi Ke Da Xue Xue Bao. 2023;43(2):242-250. [41] GRAVINA AG, PELLEGRINO R, DURANTE T, et al. The Melanocortin System in Inflammatory Bowel Diseases: Insights into Its Mechanisms and Therapeutic Potentials. Cells. 2023;12(14):1889. [42] DEBAN L, CORREALE C, VETRANO S, et al. Multiple pathogenic roles of microvasculature in inflammatory bowel disease: a Jack of all trades. Am J Pathol. 2008;172(6):1457-1466. [43] ELAHIMANESH M, NAJAFI M. Cross talk between bacterial and human gene networks enriched using ncRNAs in IBD disease. Sci Rep. 2023;13(1):7704. [44] LIU R, TANG A, WANG X, et al. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int J Mol Med. 2018;42(5):2903-2913. [45] SNEITZ N, COURT MH, ZHANG X, et al. Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics. 2009; 19(12):923-934. [46] 余水岸,李跃.粪便胆汁酸与慢性回肠末端炎症程度呈正相关:一项前瞻性队列研究[J].现代消化及介入诊疗,2023,28(10):1214-1217. [47] DUBOC H, RAJCA S, RAINTEAU D, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62(4):531-539. [48] PAN Y, ZHANG H, LI M, et al. Novel approaches in IBD therapy: targeting the gut microbiota-bile acid axis. Gut Microbes. 2024;16(1):2356284. [49] FIORUCCI S, CARINO A, BALDONI M, et al. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig Dis Sci. 2021;66(3):674-693. [50] ZHOU X, CAO L, JIANG C, et al. PPARα-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nature Commun. 2014;5(1):4573. [51] MEECH R, HU DG, MCKINNON RA, et al. The UDP-Glycosyltransferase (UGT) Superfamily: New Members, New Functions, and Novel Paradigms. Physiol Rev. 2019;99(2):1153-1222. |
| [1] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [2] | Liu Lin, Liu Shixuan, Lu Xinyue, Wang Kan. Metabolomic analysis of urine in a rat model of chronic myofascial trigger points [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1585-1592. |
| [3] | Zhao Jiacheng, Ren Shiqi, Zhu Qin, Liu Jiajia, Zhu Xiang, Yang Yang. Bioinformatics analysis of potential biomarkers for primary osteoporosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1741-1750. |
| [4] | Zhang Zhenyu, Liang Qiujian, Yang Jun, Wei Xiangyu, Jiang Jie, Huang Linke, Tan Zhen. Target of neohesperidin in treatment of osteoporosis and its effect on osteogenic differentiation of bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1437-1447. |
| [5] | Zhang Haojun, Li Hongyi, Zhang Hui, Chen Haoran, Zhang Lizhong, Geng Jie, Hou Chuandong, Yu Qi, He Peifeng, Jia Jinpeng, Lu Xuechun. Identification and drug sensitivity analysis of key molecular markers in mesenchymal cell-derived osteosarcoma [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1448-1456. |
| [6] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [7] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [8] | Ma Weibang, Xu Zhe, Yu Qiao, Ouyang Dong, Zhang Ruguo, Luo Wei, Xie Yangjiang, Liu Chen. Screening and cytological validation of cartilage degeneration-related genes in exosomes from osteoarthritis synovial fluid [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7783-7789. |
| [9] | Yan Laijun, Ge Haiya, Wang Zhengming, Yang Zongrui, Niu Lifeng, Zhan Hongsheng. Mechanism by which Tongdu Huoxue Decoction inhibits macrophage inflammation to delay intervertebral disc degeneration in rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6851-6857. |
| [10] | Nigeayi · Aihemaiti, Yilidanna · Dilixiati, An Wei, Maimaitituxun · Tuerdi. Expression of mitochondrial creatine kinase 2 in a rat model of temporomandibular joint osteoarthritis and its role in inflammation progression [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6877-6884. |
| [11] | Wang Ziheng, Wu Shuang. Oxidative stress-related genes and molecular mechanisms after spinal cord injury: data analysis and verification based on GEO database [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6893-6904. |
| [12] |
Zhou Rulin, Hu Yuanzheng, Wang Zongqing, Zhou Guoping, Zhang Baochao, Xu Qian, Bai Fanghui.
Exploration of biomarkers for moyamoya disease and analysis of traditional Chinese medicine targets#br#
#br#
[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6927-6938.
|
| [13] | Zhao Xuemei, Wang Rui, Ao · Wuliji, Bao Shuyin, Jiang Xiaohua. Effects of Agiophyllum Oligo Saccharides on inflammation and apoptosis of mouse synovial cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6939-6946. |
| [14] | Zhu Jiaping, Gao Bo, Lou Chunbiao, Yang Fengyong, Yang Kun. Monomeric traditional Chinese medicine in the treatment of rheumatoid arthritis: regulation of T cell balance [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6955-6962. |
| [15] | Yao Tingfeng, Liu Lin, Liu Shixuan, Lu Xinyue. Meta-analysis of the effectiveness of dry needling at myofascial trigger points in the treatment of knee disorders [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(32): 6989-6996. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||