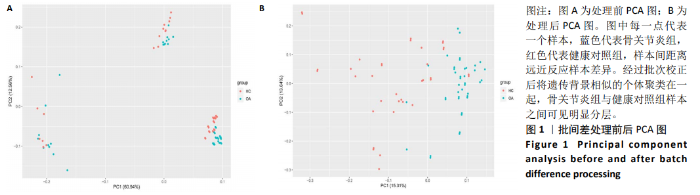
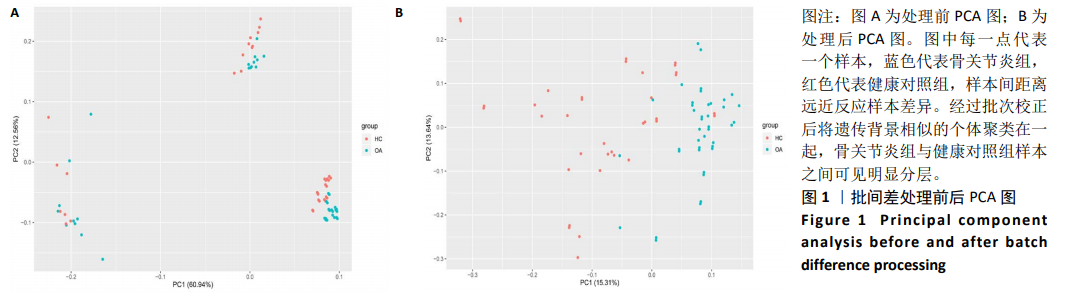
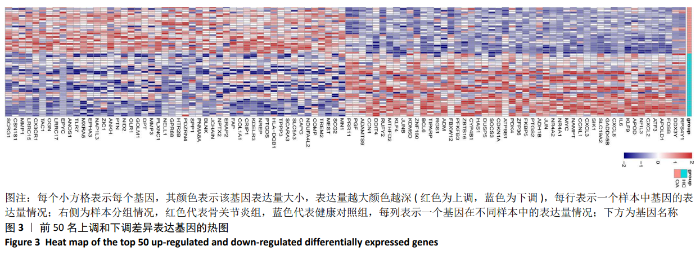
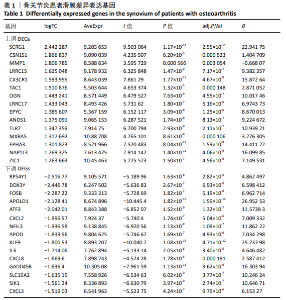
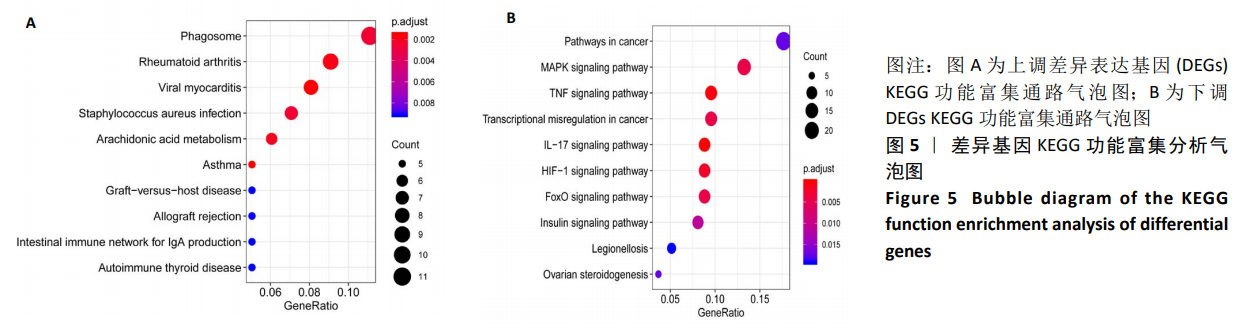
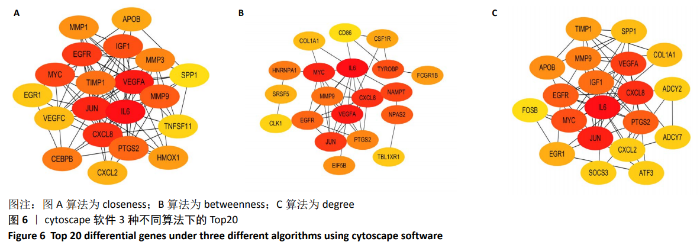
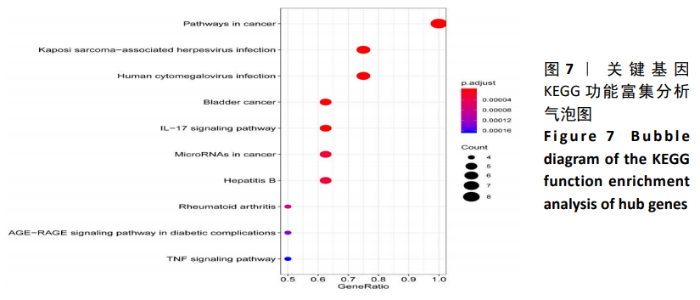
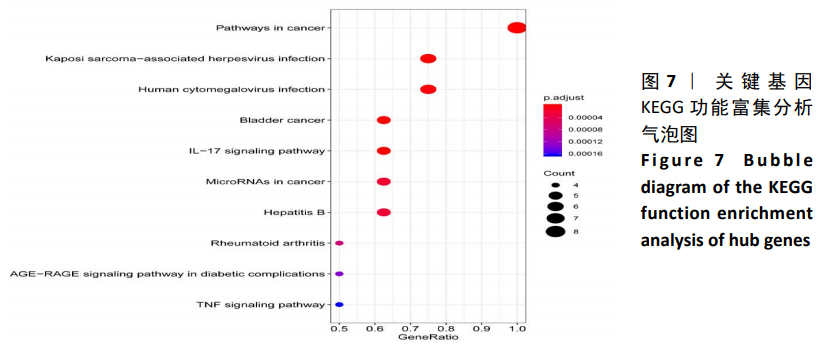
Chinese Journal of Tissue Engineering Research ›› 2021, Vol. 25 ›› Issue (5): 785-790.doi: 10.3969/j.issn.2095-4344.3016
Previous Articles Next Articles
An insight into biomarkers of osteoarthritis synovium based on bioinformatics
Song Shan, Hu Fangyuan, Qiao Jun, Wang Jia, Zhang Shengxiao, Li Xiaofeng
- Department of Rheumatology and Immunology, Second Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China
-
Received:2019-12-31Revised:2020-01-08Accepted:2020-03-18Online:2021-02-18Published:2020-12-01 -
Contact:Li Xiaofeng, Department of Rheumatology and Immunology, Second Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China -
About author:Song Shan, Department of Rheumatology and Immunology, Second Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi Province, China
CLC Number:
Cite this article
Song Shan, Hu Fangyuan, Qiao Jun, Wang Jia, Zhang Shengxiao, Li Xiaofeng. An insight into biomarkers of osteoarthritis synovium based on bioinformatics[J]. Chinese Journal of Tissue Engineering Research, 2021, 25(5): 785-790.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] SCANZELLO CR, GOLDRING SR.The role of synovitis in osteoarthritis pathogenesis.Bone. 2012; 51(2): 249-257. [2] SELLAM J, BERENBAUM F.The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol.2010;6(11): 625-635. [3] ZELZER E, OLSEN BR.Multiple roles of vascular endothelial growth factor (VEGF) in skeletal development, growth, and repair.Curr Top Dev Biol.2005;65: 169-187. [4] CHEN XY, HAO YR, WANG Z, et al.The effect of vascular endothelial growth factor on aggrecan and type II collagen expression in rat articular chondrocytes.Rheumatol Int.2012;32(11): 3359-3364. [5] ZHAO W, CAO L, YING H, et al.Endothelial CDS2 deficiency causes VEGFA-mediated vascular regression and tumor inhibition.Cell Res.2019;29(11): 895-910. [6] ZENG GQ, CHEN AB, LI W, et al.High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis. Genet Mol Res.2015;14(4): 14811-14822. [7] MEHANA EE, KHAFAGA AF, EL-BLEHI SS,The role of matrix metalloproteinases in osteoarthritis pathogenesis: An updated review.Life Sci. 2019;234: 116786. [8] KIM KS, CHOI HM, LEE YA, et al.Expression levels and association of gelatinases MMP-2 and MMP-9 and collagenases MMP-1 and MMP-13 with VEGF in synovial fluid of patients with arthritis. Rheumatol Int.2011;31(4): 543-547. [9] LERNER UH.Osteoclasts in Health and Disease. Pediatr Endocrinol Rev.2019;17(2):84-99. [10] REDLICH K, HAYER S, RICCI R, et al. Osteoclasts are essential for TNF-alpha-mediated joint destruction. J Clin Invest. 2002;110(10): 1419-1427. [11] ISLAM S, KERMODE T, SULTANA D,et al. Expression profile of protein tyrosine kinase genes in human osteoarthritis chondrocytes. Osteoarthritis Cartilage.2001;9(8): 684-693. [12] RHEE J, PARK SH, KIM SK, et al.Inhibition of BATF/JUN transcriptional activity protects against osteoarthritic cartilage destruction. Ann Rheum Dis.2017;76(2):427-434. [13] WU MH, TSAI CH, HUANG YL, et al.Visfatin Promotes IL-6 and TNF-alpha Production in Human Synovial Fibroblasts by Repressing miR-199a-5p through ERK, p38 and JNK Signaling Pathways.Int J Mol Sci. 2018;19(1). pii: E190. [14] YANG F, ZHOU S, WANG C, et al.Epigenetic modifications of interleukin-6 in synovial fibroblasts from osteoarthritis patients.Sci Rep. 2017;7: 43592. [15] MACHADO CRL, RESENDE GG, MACEDO RBV, et al.Fibroblast-like synoviocytes from fluid and synovial membrane from primary osteoarthritis demonstrate similar production of interleukin 6, and metalloproteinases 1 and 3. Clin Exp Rheumatol.2019;37(2):306-309. [16] FERNANDES MT, FERNANDES KB, MARQUEZ AS, et al.Association of interleukin-6 gene polymorphism (rs1800796) with severity and functional status of osteoarthritis in elderly individuals. Cytokine.2015;75(2):316-320. [17] RADOJČIĆ MR, THUDIUM CS, HENRIKSEN K, et al.Biomarker of extracellular matrix remodelling C1M and proinflammatory cytokine interleukin 6 are related to synovitis and pain in end-stage knee osteoarthritis patients.Pain.2017;158(7):1254-1263. [18] SHIMURA Y, KUROSAWA H, TSUCHIYA M, et al. Serum interleukin 6 levels are associated with depressive state of the patients with knee osteoarthritis irrespective of disease severity. Clin Rheumatol. 2017;36(12): 2781-2787. [19] LEE AS, ELLMAN MB, YAN D,et al.A current review of molecular mechanisms regarding osteoarthritis and pain. Gene.2013;527(2): 440-447. [20] HUANG GS,TSENG CY,LEE CH,et al.Effects of (-)-epigallocatechin-3-gallate on cyclooxygenase 2, PGE(2), and IL-8 expression induced by IL-1beta in human synovial fibroblasts.Rheumatol Int. 2010;30(9): 1197-1203. [21] YANG P, TAN J, YUAN Z, et al.Expression profile of cytokines and chemokines in osteoarthritis patients: Proinflammatory roles for CXCL8 and CXCL11 to chondrocytes.Int Immunopharmacol, 2016; 40:16-23. [22] JIA H, MA X, TONG W, et al.EGFR signaling is critical for maintaining the superficial layer of articular cartilage and preventing osteoarthritis initiation. Proc Natl Acad Sci U S A.2016; 113(50): 14360-14365. [23] JIA H, MA X, TONG W, et al EGFR Signaling: Friend or Foe for Cartilage? JBMR Plus.2019; 3(2): e10177. [24] SUN H, WU Y, PAN Z, et al.Gefitinib for Epidermal Growth Factor Receptor Activated Osteoarthritis Subpopulation Treatment.EBioMedicine.2018;32:223-233. [25] ZHANG X, ZHU J, LIU F, et al.Reduced EGFR signaling enhances cartilage destruction in a mouse osteoarthritis model. Bone Res. 2014;2:14015. |
| [1] | Min Youjiang, Yao Haihua, Sun Jie, Zhou Xuan, Yu Hang, Sun Qianpu, Hong Ensi. Effect of “three-tong acupuncture” on brain function of patients with spinal cord injury based on magnetic resonance technology [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(在线): 1-8. |
| [2] | Xu Feng, Kang Hui, Wei Tanjun, Xi Jintao. Biomechanical analysis of different fixation methods of pedicle screws for thoracolumbar fracture [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1313-1317. |
| [3] | Jiang Yong, Luo Yi, Ding Yongli, Zhou Yong, Min Li, Tang Fan, Zhang Wenli, Duan Hong, Tu Chongqi. Von Mises stress on the influence of pelvic stability by precise sacral resection and clinical validation [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1318-1323. |
| [4] | Zhang Tongtong, Wang Zhonghua, Wen Jie, Song Yuxin, Liu Lin. Application of three-dimensional printing model in surgical resection and reconstruction of cervical tumor [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1335-1339. |
| [5] | Zhang Yu, Tian Shaoqi, Zeng Guobo, Hu Chuan. Risk factors for myocardial infarction following primary total joint arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1340-1345. |
| [6] | Wei Wei, Li Jian, Huang Linhai, Lan Mindong, Lu Xianwei, Huang Shaodong. Factors affecting fall fear in the first movement of elderly patients after total knee or hip arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1351-1355. |
| [7] | Wang Jinjun, Deng Zengfa, Liu Kang, He Zhiyong, Yu Xinping, Liang Jianji, Li Chen, Guo Zhouyang. Hemostatic effect and safety of intravenous drip of tranexamic acid combined with topical application of cocktail containing tranexamic acid in total knee arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1356-1361. |
| [8] | Xiao Guoqing, Liu Xuanze, Yan Yuhao, Zhong Xihong. Influencing factors of knee flexion limitation after total knee arthroplasty with posterior stabilized prostheses [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1362-1367. |
| [9] | Peng Zhihao, Feng Zongquan, Zou Yonggen, Niu Guoqing, Wu Feng. Relationship of lower limb force line and the progression of lateral compartment arthritis after unicompartmental knee arthroplasty with mobile bearing [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1368-1374. |
| [10] | Huang Zexiao, Yang Mei, Lin Shiwei, He Heyu. Correlation between the level of serum n-3 polyunsaturated fatty acids and quadriceps weakness in the early stage after total knee arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1375-1380. |
| [11] | Zhang Chong, Liu Zhiang, Yao Shuaihui, Gao Junsheng, Jiang Yan, Zhang Lu. Safety and effectiveness of topical application of tranexamic acid to reduce drainage of elderly femoral neck fractures after total hip arthroplasty [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1381-1386. |
| [12] | Wang Haiying, Lü Bing, Li Hui, Wang Shunyi. Posterior lumbar interbody fusion for degenerative lumbar spondylolisthesis: prediction of functional prognosis of patients based on spinopelvic parameters [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1393-1397. |
| [13] | Lü Zhen, Bai Jinzhu. A prospective study on the application of staged lumbar motion chain rehabilitation based on McKenzie’s technique after lumbar percutaneous transforaminal endoscopic discectomy [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1398-1403. |
| [14] | Chen Xinmin, Li Wenbiao, Xiong Kaikai, Xiong Xiaoyan, Zheng Liqin, Li Musheng, Zheng Yongze, Lin Ziling. Type A3.3 femoral intertrochanteric fracture with augmented proximal femoral nail anti-rotation in the elderly: finite element analysis of the optimal amount of bone cement [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1404-1409. |
| [15] | Du Xiupeng, Yang Zhaohui. Effect of degree of initial deformity of impacted femoral neck fractures under 65 years of age on femoral neck shortening [J]. Chinese Journal of Tissue Engineering Research, 2021, 25(9): 1410-1416. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||