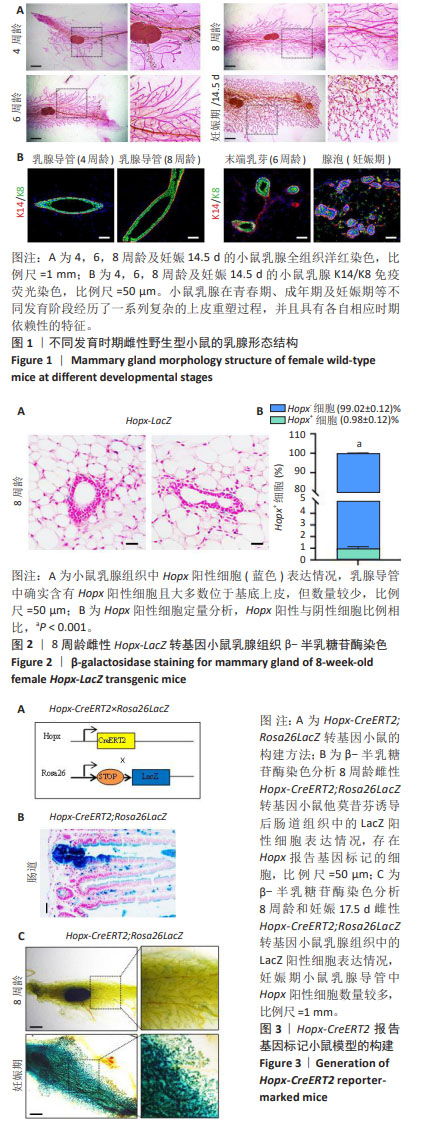
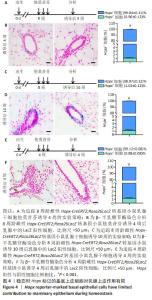
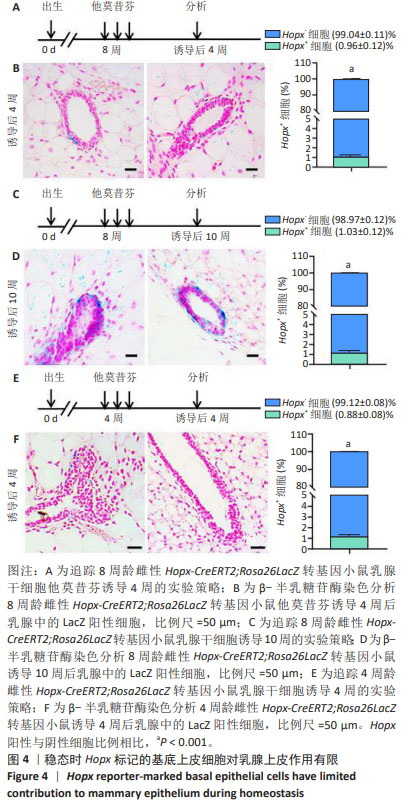
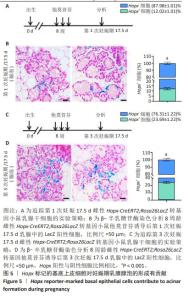
[1] SHACKLETON M, VAILLANT F, SIMPSON KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006; 439(1):84-88.
[2] ERCAN C, VAN DIEST PJ, VOOIJS M. Mammary development and breast cancer: The Role of stem cells. Curr Mol Med. 2011;11(4):270-285.
[3] FU NY, NOLAN E, LINDEMAN GJ, et al. Stem cells and the differentiation hierarchy in mammary Gland Development. Physiol Rev. 2020;100(2): 489-523.
[4] DALL G, RISBRIDGER G, BRITT K. Mammary stem cells and parity-induced breast cancer protection- new insights. J Steroid Biochem Mol Biol. 2017;170:54-60.
[5] THARMAPALAN P, MAHENDRALINGAM M, BERMAN HK, et al. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;14(38):e100852.
[6] INMAN JL, ROBERTSON C, MOTT JD, et al. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142(6):1028-1042.
[7] HENS JR, WYSOLMERSKI JJ. Key stages of mammary gland development: Molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7(5):220-224.
[8] ASSELIN-LABAT ML, VAILLANT F, SHERIDAN JM, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465(7299):798-802.
[9] CASPA GR, YAP LF, PATERSON IC. HOPX: A unique homeodomain protein in development and tumor suppression. Cancers (Basel). 2022;14(11): 1-14.
[10] ASANOMA K, KATO H, YAMAGUCHI S, et al. HOP/NECC1, a novel regulator of mouse trophoblast differentiation. J Biol Chem. 2007; 282 (33): 24065-24074.
[11] LIN CC, YAO CY, HSU YC, et al. Knock-out of Hopx disrupts stemness and quiescence of hematopoietic stem cells in mice. Oncogene. 2020;39: 5112-5123.
[12] LI D, TAKEDA N, JAIN R, et al. Hopx distinguishes hippocampal from lateral ventricle neural stem cells. Stem Cell Res. 2015;15(3):522-529.
[13] STEWART AS, SCHAAF CR, LUFF JA, et al. HOPX(+) injury-resistant intestinal stem cells drive epithelial recovery after severe intestinal ischemia. Am J Physiol Gastrointest Liver Physiol. 2021;321(5): G588-G602.
[14] TAKEDA N, JAIN R, LEBOEUF MR, et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140(8): 1655-1664.
[15] OTA C, NG-BLICHFELDT JP, KORFEI M, et al. Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Sci Rep. 2018;8(1):12983.
[16] HARADA Y, KIJIMA K, SHINMURA K, et al. Methylation of the homeobox gene, HOPX, is frequently detected in poorly differentiated colorectal cancer. Anticancer Res. 2011;31(9):2889-2892.
[17] CHEN Y, YANG L, CUI T, et al. HOPX is methylated and exerts tumour-suppressive function through Ras-induced senescence in human lung cancer. J Pathol. 2015;235(3):397-407.
[18] LEMAIRE F, MILLON R, MULLER D, et al. Loss of HOP tumour suppressor expression in head and neck squamous cell carcinoma. Br J Cancer. 2004;91(2):258-261.
[19] OOKI A, YAMASHITA K, KIKUCHI S, et al. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene. 2010;29(22):3263-3275.
[20] YAMASHITA K, KIM MS, PARK HL, et al. HOP/OB1/NECC1 promoter DNA is frequently hypermethylated and involved in tumorigenic ability in esophageal squamous cell carcinoma. Mol Cancer Res. 2008;6(1):31-41.
[21] CHEUNG WK, ZHAO M, LIU Z, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23(6):725-738.
[22] KATOH H, YAMASHITA K, WARAYA M, et al. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia. 2012;14(7):559-571.
[23] KIKUCHI M, KATOH H, WARAYA M, et al. Epigenetic silencing of HOPX contributes to cancer aggressiveness in breast cancer. Cancer Lett. 2017;384:70-78.
[24] 石建云,吴晓甜,贺 磊,等.敲减突触结合蛋白1通过抑制IL-6/STAT3信号通路改善结肠炎病变[J].中国生物化学与分子生物学报, 2022,38(10):1370-1380.
[25] GHADGE AG, DASARI P, STONE J, et al. Pubertal mammary gland development is a key determinant of adult mammographic density. Semin Cell Dev Biol. 2021;114:143-158.
[26] RICHERT MM, SCHWERTFEFEGER KL, RYDER JW, et al. An atlas of mouse mammary gland development. J Mammary Gland Bio Neoplasia. 2000;5(2):227-241.
[27] MACIAS H, HINCK L. Mammary gland development. Wiley Interdiscip Rev Dev Biol. 2012;1(4):533-557.
[28] FU NY, NOLAN E, LINDEMAN GJ, et al. Stem cells and the differentiation hierarchy in mammary gland development. Physiol Rev. 2020;100(2): 489-523.
[29] AMERONGEN RV, BOWMAN AN, NUSSE R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387-400.
[30] KEYMEULEN AV, ROCHA AS, OUSSET M, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189-193.
[31] WUIDART A, OUSSET M, RULANDS S, et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30(11):1261-1277.
[32] CHAKRABARTI R, CELIÀ-TERRASSA T, KUMAR S, et al. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science. 2018;360(6396):eaan4153.
[33] RIOS AC, FU NY, LINDEMAN GJ, et al. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506(7488):322-327.
[34] WANG DS, CAI CG, DONG XB, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015; 517(7532):81-84.
[35] FU NY, RIOS AC, PAL B, et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat Cell Biol. 2017;19(3):164-176.
[36] CAI S, KALISKY T, SAHOO D, et al. A quiescent Bcl11b high stem cell population is required for maintenance of the mammary gland. Cell Stem Cell. 2017;20(2):247-260 e245.
[37] SHIN CH, LIU ZP, ROBERT P, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110(6):725-735.
[38] FRIEDMAN CE, CHEETHAM SW, NEGI S, et al. HOPX-associated molecular programs control cardiomyocyte cell states underpinning cardiac structure and function. Dev Cell. 2024;59(1):91-107.e6.
[39] LIN CC, HSU YC, LI YH, et al. Higher HOPX expression is associated with distinct clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. Haematologica. 2017;102(6): 1044-1053.
[40] ZHAN Y, GONZALES L, VENKATADRI KOLLA, et al. Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am J Physiol Lung Cell Mol Physiol. 2006;291(2):L191-199.
[41] TAKEDA N, JAIN R, LEBOEUF MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011; 334(6061):1420-1424.
[42] TANAKA Y, KOSAKA Y, WARAYA M, et al. Differential prognostic relevance of promoter DNA methylation of CDO1 and HOPX in primary breast cancer. Anticancer Res. 2019;39(5):2289-2298.
[43] LIU C, XU Y, YANG G, et al. Niche inflammatory signals control oscillating mammary regeneration and protect stem cells from cytotoxic stress. Cell Stem Cell. 2024;31(1):89-105.e6.
[44] CHOUDHARY RK, ZHAO FQ. Stem cells in mammary health and milk production. Curr Stem Cell Res Ther. 2022;17(3):207-213.
[45] YUAN L, XIE S, BAI H, et al. Reconstruction of dynamic mammary mini gland in vitro for normal physiology and oncogenesis. Nat Methods 2023;20(12):2021-2033.
[46] BOUAMAR H, BROOME LE, LATHROP KI, et al. mTOR inhibition abrogates human mammary stem cells and early breast cancer progression markers. Breast Cancer Res. 2023;25(1):131.
|