Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (29): 6205-6211.doi: 10.12307/2025.777
Previous Articles Next Articles
Transcriptome sequencing analysis of the mechanism by which cold water swimming regulates inflammatory response in rats
Si Juncheng1, Peng Lina2, Sun Lili2, Wang Yu1, Shi Lei1, Shen Wenhui1, Li Mengqi1, Zang Wanli1
- 1Graduate School, 2School of Sports Science and Health, Harbin Sport University, Harbin 150001, Heilongjiang Province, China
-
Received:2024-07-25Accepted:2024-10-11Online:2025-10-18Published:2025-03-06 -
Contact:Peng Lina, PhD, Associate professor, Master’s supervisor, School of Sports Science and Health, Harbin Sport University, Harbin 150001, Heilongjiang Province, China -
About author:Si Juncheng, Master candidate, Graduate School, Harbin Sport University, Harbin 150001, Heilongjiang Province, China -
Supported by:Special Research Fund for Laboratory Researchers of Harbin Sport University, No. LAB2021-07 (to PLN)
CLC Number:
Cite this article
Si Juncheng, Peng Lina, Sun Lili, Wang Yu, Shi Lei, Shen Wenhui, Li Mengqi, Zang Wanli. Transcriptome sequencing analysis of the mechanism by which cold water swimming regulates inflammatory response in rats[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(29): 6205-6211.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
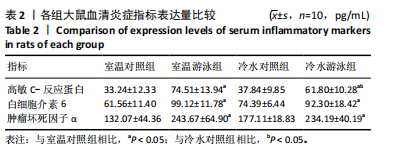
2.1 实验动物数量分析 实验过程中各组大鼠无脱失,40只大鼠全部进入结果分析。 2.2 血清高敏C-反应蛋白、白细胞介素6和肿瘤坏死因子α表达水平 如表2所示,与室温对照组相比,室温游泳组和冷水游泳组高敏C-反应蛋白、白细胞介素6和肿瘤坏死因子α表达水平显著升高(P < 0.05),冷水对照组无显著差异;与室温游泳组相比,冷水游泳组各炎症指标表达无显著差异;与冷水对照组相比,冷水游泳组白细胞介素6和肿瘤坏死因子α表达呈上升趋势,高敏C-反应蛋白表达水平显著上升(P < 0.05)。以上结果提示,温度变化对炎症反应无显著影响,游泳运动可促进炎症反应。冷水游泳组相较于室温对照组炎症反应增强,其主要影响因素可能由于运动应激。"
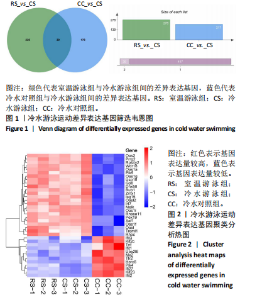
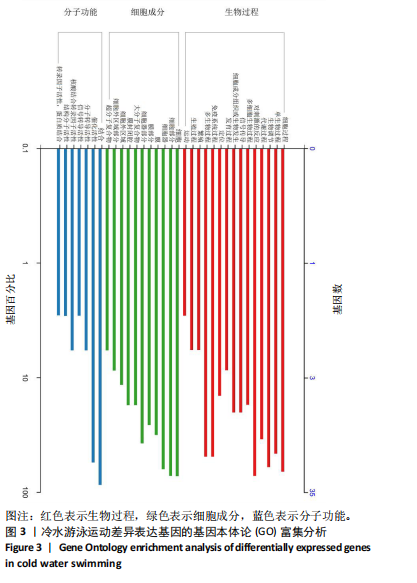
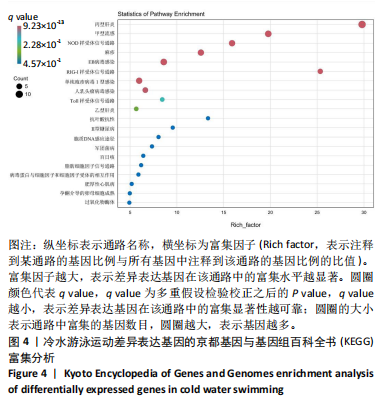
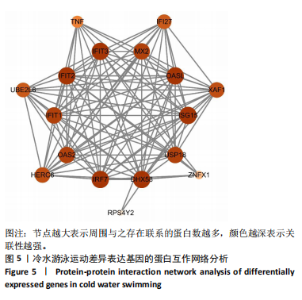
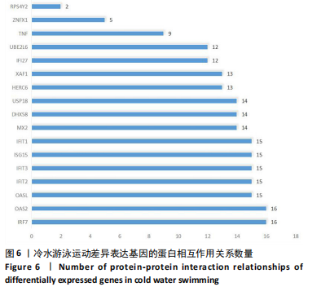
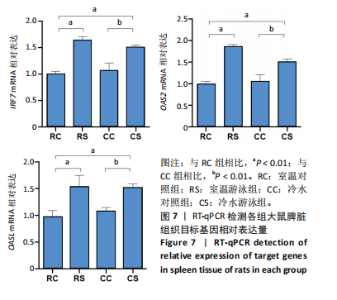
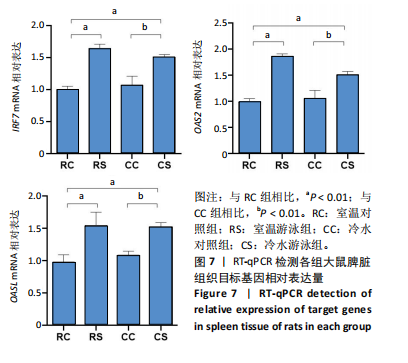
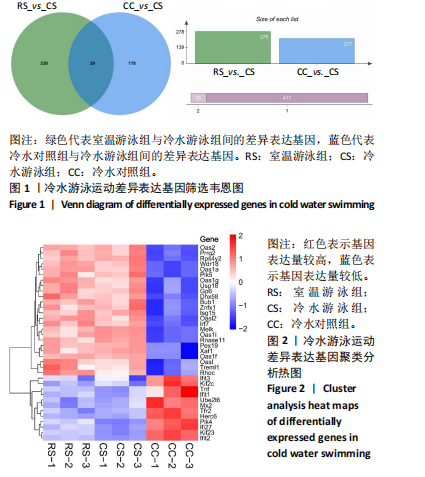
2.3 转录组分析 2.3.1 差异表达基因筛选与分析 差异表达基因筛选结果如韦恩图所示(图1)。与室温游泳组相比,冷水游泳组共有278个温度相关的差异表达基因;与冷水对照组相比,冷水游泳组共有217个游泳运动相关的差异表达基因。交集为受温度及运动干预双因素影响的差异表达基因,共39个,对该差异表达基因集进行聚类热图分析(图2)。结果显示, 各组大鼠样本重复性较好,基因相关性较强,具有可分析性。室温游泳组和冷水游泳组基因表达趋势整体相似,冷水对照组呈相反趋势,提示基因表达量差异化可能受游泳运动干预影响。 2.3.2 GO和KEGG富集分析 GO富集分析结果显示,差异表达基因富集于细胞过程、生物调节、对刺激的反应、免疫系统、运动等生物过程;参与构成细胞、细胞器、细胞膜等细胞成分;涉及结合、催化活性、分子转导活性、核酸结合转录因子活性等分子功能(图3)。KEGG富集分析结果显示,差异表达基因在丙型肝炎、甲型流感、NOD样受体信号通路、麻疹、EB病毒感染、RIG-I样受体信号通路等途径显著富集;NOD样受体信号通路富集基因数相对较多,且q value值较小,可能为关键通路,见图4。"
| [1] CHADDA KR, PUTHUCHEARY Z. Persistent inflammation, immunosuppression, and catabolism syndrome (PICS): a review of definitions, potential therapies, and research priorities. Br J Anaesth. 2024;132(3):507-518. [2] 魏伟. 炎症免疫反应软调节:药物研究新方向(英文)[J]. 中国药理学与毒理学杂志,2017,31(10):942-943. [3] 陶瑀, 谢颖, 刘秀娟. 不同运动对肥胖大鼠机体免疫和炎症反应的影响[J]. 中国体育科技,2019,55(10):74-80. [4] CERQUEIRA É, MARINHO DA, NEIVA HP, et al. Inflammatory Effects of High and Moderate Intensity Exercise-A Systematic Review. Front Physiol. 2019;10:1550. [5] 唐劲松, 宣春, 林景涛, 等. C-反应蛋白、白介素-6及降钙素原检测在新冠肺炎中的临床意义[J]. 实用医学杂志,2020,36(7):839-841. [6] HABIBI P, AHMADIASL N, NOURAZARIAN A, et al. Swimming exercise improves SIRT1, NF-κB, and IL-1β protein levels and pancreatic tissue injury in ovariectomized diabetic rats. Horm Mol Biol Clin Investig. 2022;43(3):345-352. [7] DA SILVA LA, THIRUPATHI A, COLARES MC, et al. The effectiveness of treadmill and swimming exercise in an animal model of osteoarthritis. Front Physiol. 2023;14:1101159. [8] 宋燕娟, 马春莲, 丁海超, 等. 中等强度游泳运动调控PPARγ/NF-κB/ADPN通路对2型糖尿病大鼠肝脏糖脂代谢紊乱的影响[J]. 中国康复医学杂志,2024,39(5):618-627+633. [9] COLLIER N, LOMAX M, HARPER M, et al. Habitual cold-water swimming and upper respiratory tract infection. Rhinology. 2021;59(5):485-487. [10] GIBAS-DORNA M, CHĘCIŃSKA Z, KOREK E, et al. Cold Water Swimming Beneficially Modulates Insulin Sensitivity in Middle-Aged Individuals. J Aging Phys Act. 2016;24(4):547-554. [11] CHECINSKA-MACIEJEWSKA Z, MILLER-KASPRZAK E, CHECINSKA A, et al. Gender-related effect of cold water swimming on the seasonal changes in lipid profile, ApoB/ApoA-I ratio, and homocysteine concentration in cold water swimmers. J Physiol Pharmacol. 2017;68(6):887-896. [12] DHABHAR FS. Effects of stress on immune function: the good, the bad, and the beautiful. Immunol Res. 2014;58(2-3):193-210. [13] SHATTOCK MJ, TIPTON MJ. ‘Autonomic conflict’: a different way to die during cold water immersion? J Physiol. 2012;590(14):3219-3230. [14] HOHMANN E, GLATT V, TETSWORTH K. Swimming induced pulmonary oedema in athletes - a systematic review and best evidence synthesis. BMC Sports Sci Med Rehabil. 2018;10:18. [15] STJEPANOVIC M, NIKOLAIDIS PT, KNECHTLE B. Swimming Three Ice Miles within Fifteen Hours. Chin J Physiol. 2017;60(4):197-206. [16] 潘新亮, 赵晨曦, 陈健豪, 等. 运动科学中的多组学应用前景:运动组学[J]. 北京体育大学学报,2023,46(11):52-63. [17] HSIEH YL, YANG NP, CHEN SF, et al. Early Intervention of Cold-Water Swimming on Functional Recovery and Spinal Pain Modulation Following Brachial Plexus Avulsion in Rats. Int J Mol Sci. 2022;23(3): 1178. [18] DA SILVA JT, CELLA PS, TESTA MTJ, et al. Mild-cold water swimming does not exacerbate white adipose tissue browning and brown adipose tissue activation in mice. J Physiol Biochem. 2020;76(4):663-672. [19] EN LI CHO E, ANG CZ, QUEK J, et al. Global prevalence of non-alcoholic fatty liver disease in type 2 diabetes mellitus: an updated systematic review and meta-analysis. Gut. 2023;72(11):2138-2148. [20] RIKSEN NP, BEKKERING S, MULDER WJM, et al. Trained immunity in atherosclerotic cardiovascular disease. Nat Rev Cardiol. 2023;20(12): 799-811. [21] LY M, YU GZ, MIAN A, et al. Neuroinflammation: A Modifiable Pathway Linking Obesity, Alzheimer’s disease, and Depression. Am J Geriatr Psychiatry. 2023;31(10):853-866. [22] LUO B, XIANG D, JI X, et al. The anti-inflammatory effects of exercise on autoimmune diseases: A 20-year systematic review. J Sport Health Sci. 2024;13(3):353-367. [23] 史东林, 杨贤罡. 运动员的冷环境暴露:风险因素、症状和应对策略[J]. 北京体育大学学报,2021,44(12):147-155. [24] SUGIMOTO S, MENA HA, SANSBURY BE, et al. Brown adipose tissue-derived MaR2 contributes to cold-induced resolution of inflammation. Nat Metab. 2022;4(6):775-790. [25] LIU J, WU J, QIAO C, et al. Impact of chronic cold exposure on lung inflammation, pyroptosis and oxidative stress in mice. Int Immunopharmacol. 2023;115:109590. [26] 刘萌萌, 张紫薇, 袁建彬, 等. 冷暴露对小鼠回肠机械屏障的影响及其机制[J]. 中国应用生理学杂志,2022,38(3):279-283. [27] 吴佳琦, 包雨鑫, 王梓, 等. 冷应激对科尔沁牛和科尔沁肉牛免疫功能的影响[J]. 畜牧与兽医,2021,53(12):49-52. [28] GHOLAMNEZHAD Z, SAFARIAN B, ESPARHAM A, et al. The modulatory effects of exercise on lipopolysaccharide-induced lung inflammation and injury: A systemic review. Life Sci. 2022;293:120306. [29] ZHANG Y, XU J, ZHOU D, et al. Swimming exercise ameliorates insulin resistance and nonalcoholic fatty liver by negatively regulating PPARγ transcriptional network in mice fed high fat diet. Mol Med. 2023;29(1): 150. [30] 李章春, 孙海波, 秦萍, 等. 游泳运动对慢性非细菌性前列腺炎大鼠p38MAPK/NF-κB信号通路的影响[J]. 医学理论与实践,2024, 37(13):2165-2167+2180. [31] ALLEN J, SUN Y, WOODS JA. Exercise and the Regulation of Inflammatory Responses. J Prog Mol Biol Transl Sci. 2015;135:337-354. [32] ALMURAIKHY S, DOUDIN A, DOMLING A, et al. Molecular regulators of exercise-mediated insulin sensitivity in non-obese individuals. J Cell Mol Med. 2024;28(1):e18015. [33] SMITH LL. Cytokine hypothesis of overtraining: a physiological adaptation to excessive stress? Med Sci Sports Exerc. 2000;32(2): 317-331. [34] LIN W, SONG H, SHEN J, et al. Functional role of skeletal muscle-derived interleukin-6 and its effects on lipid metabolism. Front Physiol. 2023; 14:1110926. [35] JIANG S, BAE JH, WANG Y, et al. The Potential Roles of Myokines in Adipose Tissue Metabolism with Exercise and Cold Exposure. Int J Mol Sci. 2022;23(19):11523. [36] MARTINS GA, DEGEN AN, ANTUNES FTT, et al. Benefits of electroacupuncture and a swimming association when compared with isolated protocols in an osteoarthritis model. J Tradit Complement Med. 2022;12(4):375-383. [37] BOSIACKI M, TARNOWSKI M, MISIAKIEWICZ-HAS K, et al. The Effect of Cold-Water Swimming on Energy Metabolism, Dynamics, and Mitochondrial Biogenesis in the Muscles of Aging Rats. Int J Mol Sci. 2024;25(7):4055. [38] 王寅彪, 冯继伟, 陈礼朋, 等. 2′-5′寡聚腺苷酸合成酶的抗病毒作用及机制研究进展[J]. 新乡医学院学报,2021,38(5):485-487. [39] LIU M, ZHANG Y, YAN J, et al. Aerobic exercise alleviates ventilator-induced lung injury by inhibiting NLRP3 inflammasome activation. BMC Anesthesiol. 2022;22(1):369. [40] XU Z, MA Z, ZHAO X, et al. Aerobic exercise mitigates high-fat diet-induced cardiac dysfunction, pyroptosis, and inflammation by inhibiting STING-NLRP3 signaling pathway. Mol Cell Biochem. 2024;479(12): 3459-3470. [41] 耿元文, 林琴琴, 李若明, 等. 一次性力竭运动模型促进大鼠肾脏NOD样受体蛋白3炎性小体的表达[J]. 中国组织工程研究,2022, 26(2):190-196. [42] 高晴, 鱼文欢, 何佳静, 等. 核苷酸结合寡聚化结构域样受体蛋白3炎症小体在PCOS中的研究进展[J]. 生殖医学杂志,2022,31(3): 420-424. |
| [1] | De Ji, Suo Langda, Wei Yuchen, Wang Bin, Awangcuoji, Renqingcuomu, Cui Jiuzeng, Zhang Lei, Ba Gui. Comprehensive analysis of genes related to endometrial receptivity and alternative splicing events in northwest Tibetan cashmere goats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1429-1436. |
| [2] | He Bo, Chen Wen, Ma Suilu, He Zhijun, Song Yuan, Li Jinpeng, Liu Tao, Wei Xiaotao, Wang Weiwei, Xie Jing . Pathogenesis and treatment progress of flap ischemia-reperfusion injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1230-1238. |
| [3] | He Yang, Tang Buyuan, Lu Changhuai. Molecular mechanisms of ligament flavum hypertrophy: analysis based on methylation sequencing and transcriptome integration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1013-1020. |
| [4] | Yu Qinghe, Cai Ziming, Wu Jintao, Ma Pengfei, Zhang Xin, Zhou Longqian, Wang Yakun, Lin Xiaoqin, Lin Wenping. Vanillic acid inhibits inflammatory response and extracellular matrix degradation of endplate chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6391-9397. |
| [5] | Fan Jiaxin, Jia Xiang, Xu Tianjie, Liu Kainan, Guo Xiaoling, Zhang Hui, Wang Qian . Metformin inhibits ferroptosis and improves cartilage damage in osteoarthritis model rats [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6398-6408. |
| [6] | Zhou Ying, Tian Yong, Zhong Zhimei, Gu Yongxiang, Fang Hao. Inhibition of tumor necrosis factor receptor associated factor 6 regulates mTORC1/ULK1 signaling and promotes autophagy to improve myocardial injury in sepsis mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6434-6440. |
| [7] | Wang Wanchun, , Yi Jun, Yan Zhangren, Yang Yue, Dong Degang, Li Yumei. 717 Jiedu Decoction remodels homeostasis of extracellular matrix and promotes repair of local injured tissues in rats after Agkistrodon halys bite [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6457-6465. |
| [8] | Zhang Xin, Guo Baojuan, Xu Huixin, Shen Yuzhen, Yang Xiaofan, Yang Xufang, Chen Pei. Protective effects and mechanisms of 3-N-butylphthalide in Parkinson’s disease cell models [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6466-6473. |
| [9] | Zhang Songjiang, Li Longyang, Zhou Chunguang, Gao Jianfeng. Central anti-inflammatory effect and mechanism of tea polyphenols in exercise fatigue model mice [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6474-6481. |
| [10] | Hu Shujuan, Liu Dang, Ding Yiting, Liu Xuan, Xia Ruohan, Wang Xianwang. Ameliorative effect of walnut oil and peanut oil on atherosclerosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6482-6488. |
| [11] | Zhang Jian, Cai Feng, Li Tingwen, Ren Pengbo. Fatigue gait recognition of athletes based on fish swarm algorithm [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6489-6498. |
| [12] | Zhang Zihan¹, Wang Jiaxin¹, Yang Wenyi², Zhu Lei¹. Regulatory mechanism of exercise promoting mitochondrial biogenesis in skeletal muscle [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6499-6508. |
| [13] | Wang Jianxu, Dong Zihao, Huang Zishuai, Li Siying, Yang Guang. Interaction between immune microenvironment and bone aging and treatment strategies [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6509-6519. |
| [14] | Zhang Bochun, Li Wei, Li Guangzheng, Ding Haoqin, Li Gang, Liang Xuezhen, . Association between neuroimaging changes and osteonecrosis: a large sample analysis from UK Biobank and FinnGen databases [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6574-6582. |
| [15] | Xu Zhi, Chen Yundong, Sun Yujie, Gong Xiaonan, Li Yuwan. Data of spinal osteosarcoma patients in United States based on SEER database: construction and validation of a prediction model for treatment outcomes and prognosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(30): 6583-6590. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||