Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (1): 193-201.doi: 10.12307/2025.003
Previous Articles Next Articles
Application of single-cell RNA sequencing technology in Parkinson’s disease
Liu Ziyu1, Geng Dandan1, 2, Zhang Runjiao1, Liu Qing1, Li Yibo1, Wang Hongfang1, Xie Wenmeng1, Wang Wenyu1, Hao Jiaxin1, Wang Lei1, 2
- 1Department of Human Anatomy and Histoembryology, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China; 2Key Laboratory of Neural and Vascular Biology, Ministry of Education, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China
-
Received:2023-10-30Accepted:2023-12-14Online:2025-01-08Published:2024-05-20 -
Contact:Wang Lei, PhD, Professor, Doctoral supervisor, Department of Human Anatomy and Histoembryology, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China; Key Laboratory of Neural and Vascular Biology, Ministry of Education, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China -
About author:Liu Ziyu, Master candidate, Department of Human Anatomy and Histoembryology, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China -
Supported by:Wang Lei, PhD, Professor, Doctoral supervisor, Department of Human Anatomy and Histoembryology, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China; Key Laboratory of Neural and Vascular Biology, Ministry of Education, Hebei Medical University, Shijiazhuang 050017, Hebei Province, China
CLC Number:
Cite this article
Liu Ziyu, Geng Dandan, Zhang Runjiao, Liu Qing, Li Yibo, Wang Hongfang, Xie Wenmeng, Wang Wenyu, Hao Jiaxin, Wang Lei. Application of single-cell RNA sequencing technology in Parkinson’s disease[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(1): 193-201.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

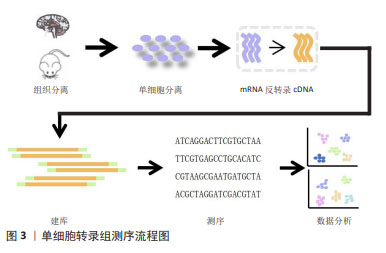
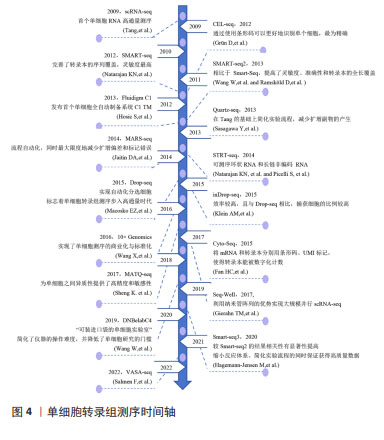
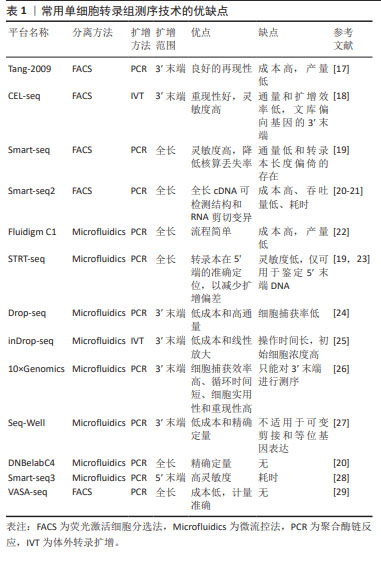
2.1.1 单细胞或单细胞核的分离 单细胞或单细胞核的分离和捕获是从组织中获得高质量单个细胞的过程,为提取精确的遗传和生化信息提供条件。低通量的单细胞分离传统方法包括连续稀释法、显微操作、激光捕获显微切割等,这类传统的方法依赖于人工在高倍显微镜下观察细胞的形态和颜色等特征来挑取单个细胞,要求操作者技术熟练,具有难度较大、产出率较低等明显缺陷。常用的高通量方法包括荧光激活细胞分选法和微流控法[12-15]。 其中荧光激活细胞分选法根据细胞的特征进行快速分离,具有灵敏度高、准确度高、通量高等优点,但需要细胞初始样本量较大。目前常用的是微流控技术,包括液滴式微流控和芯片式微流控2种方式,将细胞分离至nL级的微容器中进行捕获单个细胞,具有高分辨率、高灵敏度和所需求样本及试剂少等特点,是目前单细胞分离最好的方法。市场上比较成熟的商业scRNA-seq分离方法主要是10X Genomics公司的Chromium(液滴法)及BD公司的Rhapsody(微孔法),都是集细胞分选、细胞溶解、标记建库于一体的方法。 2.1.2 反转录和cDNA的扩增 scRNA-seq的准确性和灵敏度取决于其在反转录和扩增过程中的效率,从单个细胞有限的mRNA中获得足够的cDNA,从而能够在单细胞水平上进行转录组学分析。单细胞中的遗传物质含量非常小(通常< 10 pg/细胞),无法满足进一步进行二代测序(200 ng以上)的要求,因此需对单细胞测序样本进行扩增。在将mRNA转化为第一链cDNA之后,得到的cDNA进行聚合酶链反应(polymerase chain reaction,PCR)或体外转录扩增 (in vitro transcription,IVT)[9,16]。 PCR作为一种非线性扩增过程应用于Smart-seq,Smart-seq2,Fluidigm C1,Drop-seq和10x Genomics。体外转录扩增是另一种扩增方法和线性扩增过程,用于CEL-seq,MARS-Seq和Drop-seq。它需要对扩增的RNA进行额外一轮的反转录,这会导致额外的3’覆盖偏差,这2种方法都可能导致放大偏差。为了克服扩增相关偏差,引入了唯一分子标识符,在反转录步骤中对细胞内每个单独的mRNA分子进行条形码编码,从而提高scRNA-seq的定量能力并通过消除PCR扩增偏差来提高读取准确性。见表1。 2.1.3 构建测序文库 对扩增片段进行质检,质检合格后建库。文库制备包括将扩增产物随机打断成小DNA片段、末端修复、添加接头和PCR扩增,从而得到所需文库。测序技术主要分为全长(Full-length)测序与基于标签(Tag-based)测序[30-32]。前者方法比较简单直接,但也存在明显缺陷:低通量、高成本,当所测细胞数量较低时,研究结果不足以反映真实问题,并且随着所测细胞数量增多,测序成本也随之呈线性提升。而新进的基于Tag-based的单细胞识别,即给每个不同的细胞赋予一段独一无二的DNA序列,混合测序时可把携带相同条形码编码序列的核酸分子视为来自同一细胞,从而可通过一次建库,测得成百上千个单细胞信息并加以区分。显然,此种方法相较于传统方法优势更大。 "

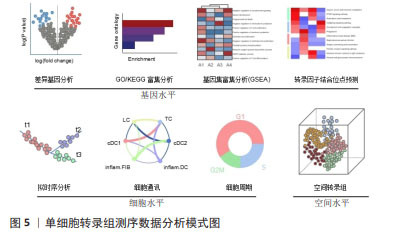
2.1.4 数据处理和分析 测序的原始数据首先需要生成FASTQ reads格式,之后进行质量控制,基于每个样本检测到的唯一分子标识符、基因、线粒体等的分布进行统计,从而计算出所需过滤阈值,过滤掉不合格的数据后再转换成基因表达矩阵进行基础信息分析。根据每个基因的分散度,对差异基因进行鉴定,随后将整合分析后的数据进行主成分分析(principal component analysis,PCA)、T-分布随机邻域嵌入(T-distributed stochastic neighbor embedding,t-SNE)和统一流形逼近与投影(uniform manifold approximation and projection,UMAP)等可视化降维分析。对细胞进行自动聚类分析后,根据软件及相关文献选择每个Cluster特异且高表达的基因,作为其标记基因进行注释。除鉴定细胞类型,得到的数据还可进行拟时序分析、调控网络分析、生物信息学分析等,深入挖掘细胞状态转换、细胞功能和相互作用机制[33-36],见图5。"

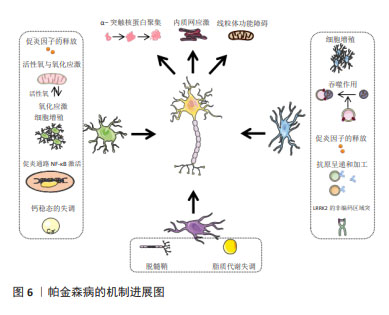
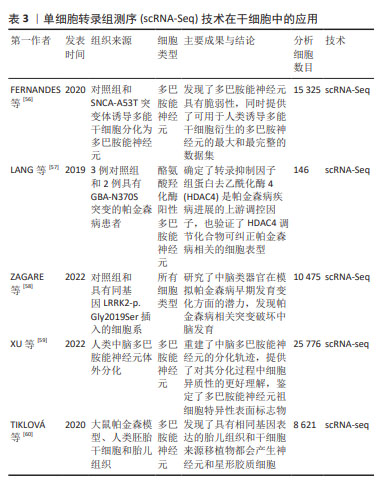
2.2.1 分析DaNs DaNs主要位于中脑的黑质和腹侧被盖区,可投射至其他脑区用以调节其他神经元活动。DaNs释放的多巴胺是大脑中必不可少的神经递质,在调节运动、认知、记忆、奖励等方面起着关键作用。DaNs在帕金森病患者中逐渐退化,特别是尾状壳核中DaNs的耗竭已被证明是帕金森病运动症状的基础[45-46],然而,有研究也发现黑质致密部中一些DaNs甚至可以存活到帕金森病的晚期阶段,这表明不同的DaNs亚型对帕金森病导致的神经退化具有差异性,即脆弱性[47]。因此,DaNs的亚型和分子特性引起了人们极大的兴趣,对于它们如何参与帕金森病的发病需要深入研究。 DaNs的易感性和脆弱性是引起帕金森病的重要因素之一,通过scRNA-seq技术全面了解DaNs及其在帕金森病中的脆弱性,对于研究帕金森病相关发病机制是至关重要的。研究团队为探索帕金森病转录组学的早期失调[42],利用snRNA-seq对6月龄帕金森病模型小鼠(PDGF-hα- Synuclein A53T,α-syn-A53T)及野生型小鼠中脑进行测序,发现DaNs中钙电压门控通道亚基α1 A(Calcium voltage- gated channel subunit alpha1 A,Cacna1a)和钙电压门控通道亚基α1 B(Calcium voltage-gated channel subunit alpha1 B,Cacna1b)通过诱导钙电流释放神经递质的能力缺陷,这可能导致多巴胺水平降低和多巴胺能轴突投射的生成过程受损。POULIN等[37]将1-甲基-4-苯基-1,2,3,6-四氢吡啶(1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine,MPTP)制备的帕金森病模型小鼠DaNs进行scRNA-seq,发现共表达SRY盒转录因子6(SRY-Box transcription factor 6,Sox6)和醛脱氢酶1家族成员A1(aldehyde dehydrogenase 1 family member A1,Aldh1a1)的腹侧DaNs对神经毒素MPTP更具脆弱性。正如先前的报道,在帕金森病中位于腹侧黑质致密部的DaNs比腹侧被盖区更脆弱。帕金森病发生运动障碍的原因可能是位于腹侧黑质致密部的DaNs通过黑质-纹状体通路支配背外侧纹状体,严重影响帕金森病的运动功能[48]。此外,KRAMER等[39]对6-羟多巴胺模型小鼠的DaNs进行了scRNA-seq,发现位于腹侧被盖区高表达神经源性分化蛋白6(monoclonal antibody to neurogenic differentiation 6,Neurod6)的DaNs在帕金森病模型中优先保留,这与Neurod6的潜在神经保护作用一致。因此,scRNA-seq有助于更清楚地了解脆弱性神经元的特定分子特征,为完善帕金森病的实验室模型、改善帕金森病的预后或进展以及特异性细胞类型的治疗提供机会。 在帕金森病小鼠模型中取得了重大进展的同时,基于scRNA-seq技术也对成人脑神经元的部分脑区进行全面分类。2016年LAKE等[38]对正常女性的6个大脑新皮质区域进行基因表达聚类,定义了16个神经元亚型。通过对这些神经元细胞亚型的认知,来构建一个完整的体系,这为广泛采集人类大脑表达基因打开了大门。随后,AGARWAL 等[49]对5例帕金森病患者和健康正常人的黑质和大脑皮质区域进行了snRNA-seq,通过分析人类黑质转录组细胞图谱中特异性基因表达来识别特定的细胞类型,首次在人类中得到帕金森病的常见遗传风险与DaNs特异性表达基因之间存在显著关联,同时也描绘出第一个全面的人类黑质细胞类型图谱。另外,KAMATH等[44]通过snRNA-seq技术对帕金森病患者和匹配对照的黑质致密部DaNs分析,发现共表达SOX6和血管紧张素Ⅱ受体1(angiotensin II Receptor type 1,AGTR1)的DaNs亚群更容易受到帕金森病神经变性的影响,并且这些神经元富集于线粒体功能障碍、运动神经元死亡、氧化应激等功能。遗憾的是,与小鼠相比,scRNA-seq技术对人类黑质的研究匮乏,还需要进行更加深入的研究。 2.2.2 分析神经胶质细胞 虽然DaNs的丢失是帕金森病病理学的关键基础,但已有证据表明胶质细胞在神经退行性疾病的发生发展中起着必不可少的作用[50]。神经胶质细胞主要包括星形胶质细胞、小胶质细胞和少突胶质细胞,它们可以调节神经元活动且不产生电脉冲,被认为是神经元的支持细胞且在细胞多样性和功能上优于神经元[51]。利用scRNA-seq分析显著提高了对帕金森病中胶质细胞反应的了解,从而鉴定出小鼠模型和人类样本中与神经变性相关的特殊胶质细胞亚群,为进一步探究帕金森病的发病机制提供帮助。 帕金森病的研究虽然主要集中在黑质DaNs上,但最近scRNA-seq研究结果表明,非神经元和黑质以外的大脑区域也发生了与疾病相关的变化。ZHONG等[42]对6月龄帕金森病模型小鼠的中脑、纹状体、小脑组织进行snRNA-seq,发现了多种细胞类型转录水平的改变,包括蛋白稳态、离子通道、细胞外环境和神经炎症等方面具有明显的功能障碍。其中中脑和纹状体中的星形胶质细胞和少突胶质细胞表现出帕金森病风险基因的强烈富集、核因子κB活性的升高、离子通道成分的改变、蛋白质稳态的失衡以及谷氨酸能信号传导的失调。GUO等[52]通过snRNA-Seq技术揭示了帕金森病模型小鼠大脑皮质、海马体、纹状体和小脑4个脑区中疾病特异性细胞状态的变化,并首次揭示了疾病相关的星形胶质细胞AST1亚群从静止状态到过度激活状态的转录调控,转录因子Dbx2和Sox13被确定为帕金森病模型中与AST1亚群最特异性相关的转录因子,这为治疗帕金森病提供了潜在的靶标标志物。随后,SCHONHOFF等[41]构建AAV2-SYN腺相关病毒通过脑立体定向注射到黑质致密部构建帕金森病小鼠模型,通过scRNA-Seq发现与疾病相关小胶质细胞亚群的GO功能富集于Toll样受体信号传导、吞噬作用、抗原呈递和加工以及细胞因子、趋化因子信号传导等途径。这些对帕金森病模型小鼠的研究为理解疾病发病机制背后的细胞异质性提供了参考。 在帕金森病模型小鼠中对胶质细胞的作用机制进行实验研究的同时,在人类样本中也进行了相关探索。有研究对人类黑质和大脑皮质进行了snRNA-seq[49],鉴定出所有主要细胞类型,其中胶质细胞占95.5%,包括星形胶质细胞、小胶质细胞、少突胶质细胞、少突胶质前体细胞、壁细胞(内皮细胞和周细胞)等,少突胶质细胞占胶质细胞的72%。研究发现,帕金森病中常见的遗传风险除了与DaNs特异基因表达相关以外,也与少突胶质细胞特异基因表达存在明显关联,这些基因主要参与代谢过程、基因调控、激酶活性、蛋白磷酸化和神经发生等过程[53]。另有研究对6例原发性帕金森病患者和5例性别和年龄相匹配的对照者尸检脑组织进行snRNA-seq分析[43],发现帕金森病患者中脑黑质中星形胶质细胞和小胶质细胞数目增加,差异基因主要富集于细胞增殖、未折叠蛋白反应、细胞因子信号传导等信号通路,表明星形胶质细胞和小胶质细胞处于激活状态;少突胶质细胞数量减少,并且星形胶质细胞和小胶质细胞的增多与少突胶质细胞减少部分相吻合;其余细胞表现为应激诱导的S100钙结合蛋白上调。LANGSTON等[54]将人类额叶皮质进行snRNA-seq分析,同时取该患者外周血单核细胞,诱导成多能干细胞分化为小胶质细胞(iMicroglia)进行scRNA-seq,并将皮质小胶质细胞的测序结果与iMicroglia进行比较,发现帕金森病发病风险是由小胶质细胞中LRRK2的非编码区域突变所介导。综合以上研究结果,scRNA-seq为明确中脑神经胶质细胞对帕金森病的潜在贡献提供重要见解。 2.3 scRNA-seq有助于临床治疗 截至今天,帕金森病常见的治疗方法主要是对症治疗,包括以左旋多巴为代表的药物治疗、康复技术加以辅助以及手术治疗如脑立体定向手术、深部脑刺激术等[55]。但这些治疗只能缓解一些症状,并不会减缓疾病的进展,且长期疗效不明。药物在控制该疾病的同时会产生中枢性和外周性两类不良反应,外周性不良反应会随着治疗的进展以及人体逐渐适应药物,症状逐渐消失;中枢性不良反应症状与帕金森病原发症状存在高度的相似性,如非自主性晃动、运动障碍等,严重影响患者的生活质量。因此,scRNA-seq被用于揭示帕金森病新的发病机制和提供有效的治疗靶点。 见表3。 "
| [1] YE H, ROBAK LA, YU M, et al. Genetics and Pathogenesis of Parkinson’s Syndrome. Annu Rev Pathol. 2023;18:95-121. [2] HAYES MT. Parkinson’s Disease and Parkinsonism. Am J Med. 2019;132(7): 802-807. [3] BLOEM BR, OKUN MS, KLEIN C. Parkinson’s disease. Lancet. 2021;397 (10291):2284-2303. [4] PAJARES M, I ROJO A, MANDA G, et al. Inflammation in Parkinson’s Disease: Mechanisms and Therapeutic Implications. Cells. 2020;9(7):1687. [5] ZHANG S, LIU YQ, JIA C, et al. Mechanistic basis for receptor-mediated pathological α-synuclein fibril cell-to-cell transmission in Parkinson’s disease. Proc Natl Acad Sci U S A. 2021;118(26):e2011196118. [6] BAE YJ, KIM JM, SOHN CH, et al. Imaging the Substantia Nigra in Parkinson Disease and Other Parkinsonian Syndromes. Radiology. 2021;300(2): 260-278. [7] VRIJSEN S, HOUDOU M, CASCALHO A, et al. Polyamines in Parkinson’s Disease: Balancing Between Neurotoxicity and Neuroprotection. Annu Rev Biochem. 2023;92:435-464. [8] TANG F, BARBACIORU C, WANG Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377-382. [9] HEDLUND E, DENG Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol Aspects Med. 2018;59:36-46. [10] PAPALEXI E, SATIJA R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35-45. [11] LI X, WANG CY. From bulk, single-cell to spatial RNA sequencing. Int J Oral Sci. 2021;13(1):36. [12] HWANG B, LEE JH, BANG D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50(8):1-14. [13] GROSS A, SCHOENDUBE J, ZIMMERMANN S, et al. Technologies for Single-Cell Isolation. Int J Mol Sci. 2015;16(8):16897-16919. [14] STUART T, SATIJA R. Integrative single-cell analysis. Nat Rev Genet. 2019; 20(5):257-272. [15] PAN Y, CAO W, MU Y, et al. Microfluidics Facilitates the Development of Single-Cell RNA Sequencing. Biosensors (Basel). 2022;12(7):450. [16] KISELEV VY, ANDREWS TS, HEMBERG M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20(5):273-282. [17] WU X, LIU T, YE C, et al. scAPAtrap: identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data. Brief Bioinform. 2021;22(4):bbaa273. [18] GRÜN D, KESTER L, VAN OUDENAARDEN A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11(6):637-640. [19] NATARAJAN KN. Single-Cell Tagged Reverse Transcription (STRT-Seq). Methods Mol Biol. 2019;1979:133-153. [20] WANG W, ZHONG Y, ZHUANG Z, et al. Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer. Clin Transl Med. 2021;11(1):e253. [21] RAMSKÖLD D, LUO S, WANG YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777-782. [22] HOSIC S, MURTHY SK, KOPPES AN. Microfluidic Sample Preparation for Single Cell Analysis. Anal Chem. 2016;88(1):354-380. [23] PICELLI S, FARIDANI OR, BJÖRKLUND AK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171-181. [24] MACOSKO EZ, BASU A, SATIJA R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015; 161(5):1202-1214. [25] KLEIN AM, MAZUTIS L, AKARTUNA I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187-1201. [26] WANG X, HE Y, ZHANG Q, et al. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Genomics Proteomics Bioinformatics. 2021;19(2):253-266. [27] GIERAHN TM, WADSWORTH MH 2ND, HUGHES TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14(4):395-398. [28] HAGEMANN-JENSEN M, ZIEGENHAIN C, CHEN P, et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol. 2020;38(6):708-714. [29] SALMEN F, DE JONGHE J, KAMINSKI TS, et al. High-throughput total RNA sequencing in single cells using VASA-seq. Nat Biotechnol. 2022;40(12): 1780-1793. [30] JOVIC D, LIANG X, ZENG H, et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med. 2022;12(3):e694. [31] HONG M, TAO S, ZHANG L, et al. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol. 2020;13(1):166. [32] WU Y, ZHANG K. Tools for the analysis of high-dimensional single-cell RNA sequencing data. Nat Rev Nephrol. 2020;16(7):408-421. [33] ZHAO T, FU Y, ZHU J, et al. Single-Cell RNA-Seq Reveals Dynamic Early Embryonic-like Programs during Chemical Reprogramming. Cell Stem Cell. 2018;23(1):31-45.e7. [34] BALZER MS, MA Z, ZHOU J, et al. How to Get Started with Single Cell RNA Sequencing Data Analysis. J Am Soc Nephrol. 2021;32(6):1279-1292. [35] HUANG M, XU L, LIU J, et al. Cell-Cell Communication Alterations via Intercellular Signaling Pathways in Substantia Nigra of Parkinson’s Disease. Front Aging Neurosci. 2022;14:828457. [36] YIP SH, SHAM PC, WANG J. Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Brief Bioinform. 2019;20(4):1583-1589. [37] POULIN JF, ZOU J, DROUIN-OUELLET J, et al. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep. 2014; 9(3):930-943. [38] LAKE BB, AI R, KAESER GE, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586-1590. [39] KRAMER DJ, RISSO D, KOSILLO P, et al. Combinatorial Expression of Grp and Neurod6 Defines Dopamine Neuron Populations with Distinct Projection Patterns and Disease Vulnerability. eNeuro. 2018;5(3): ENEURO.0152-18.2018. [40] WELCH JD, KOZAREVA V, FERREIRA A, et al. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell. 2019;177(7):1873-1887.e17. [41] SCHONHOFF AM, FIGGE DA, WILLIAMS GP, et al. Border-associated macrophages mediate the neuroinflammatory response in an alpha-synuclein model of Parkinson disease. Nat Commun. 2023;14(1):3754. [42] ZHONG J, TANG G, ZHU J, et al. Single-cell brain atlas of Parkinson’s disease mouse model. J Genet Genomics. 2021;48(4):277-288. [43] SMAJIĆ S, PRADA-MEDINA CA, LANDOULSI Z, et al. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain. 2022;145(3):964-978. [44] KAMATH T, ABDULRAOUF A, BURRIS SJ, et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson’s disease. Nat Neurosci. 2022; 25(5):588-595. [45] TIKLOVÁ K, BJÖRKLUND ÅK, LAHTI L, et al. Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat Commun. 2019;10(1):581. [46] HOOK PW, MCCLYMONT SA, CANNON GH, et al. Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am J Hum Genet. 2018;102(3):427-446. [47] MASATO A, PLOTEGHER N, BOASSA D, et al. Impaired dopamine metabolism in Parkinson’s disease pathogenesis. Mol Neurodegener. 2019;14(1):35. [48] GUATTEO E, BERRETTA N, MONDA V, et al. Pathophysiological Features of Nigral Dopaminergic Neurons in Animal Models of Parkinson’s Disease. Int J Mol Sci. 2022;23(9):4508. [49] AGARWAL D, SANDOR C, VOLPATO V, et al. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun. 2020;11(1):4183. [50] WANG ZL, YUAN L, LI W, et al. Ferroptosis in Parkinson’s disease: glia-neuron crosstalk. Trends Mol Med. 2022;28(4):258-269. [51] COPE EC, GOULD E. Adult Neurogenesis, Glia, and the Extracellular Matrix. Cell Stem Cell. 2019;24(5):690-705. [52] GUO Y, MA J, HUANG H, et al. Defining Specific Cell States of MPTP-Induced Parkinson’s Disease by Single-Nucleus RNA Sequencing. Int J Mol Sci. 2022; 23(18):10774. [53] BRYOIS J, SKENE NG, HANSEN TF, et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat Genet. 2020;52(5):482-493. [54] LANGSTON RG, BEILINA A, REED X, et al. Association of a common genetic variant with Parkinson’s disease is mediated by microglia. Sci Transl Med. 2022;14(655):eabp8869. [55] ARMSTRONG MJ, OKUN MS. Choosing a Parkinson Disease Treatment. JAMA. 2020;323(14):1420. [56] FERNANDES HJR, PATIKAS N, FOSKOLOU S, et al. Single-Cell Transcriptomics of Parkinson’s Disease Human In Vitro Models Reveals Dopamine Neuron-Specific Stress Responses. Cell Rep. 2020;33(2):108263. [57] LANG C, CAMPBELL KR, RYAN BJ, et al. Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifies HDAC4 as a Regulator of Parkinson Cell Phenotypes. Cell Stem Cell. 2019;24(1):93-106.e6. [58] ZAGARE A, BARMPA K, SMAJIC S, et al. Midbrain organoids mimic early embryonic neurodevelopment and recapitulate LRRK2-p.Gly2019Ser-associated gene expression. Am J Hum Genet. 2022;109(2):311-327. [59] XU P, HE H, GAO Q, et al. Human midbrain dopaminergic neuronal differentiation markers predict cell therapy outcomes in a Parkinson’s disease model. J Clin Invest. 2022;132(14):e156768. [60] TIKLOVÁ K, NOLBRANT S, FIORENZANO A, et al. Single cell transcriptomics identifies stem cell-derived graft composition in a model of Parkinson’s disease. Nat Commun. 2020;11(1):2434. [61] BARKER RA, GÖTZ M, PARMAR M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557(7705):329-334. [62] FIORENZANO A, SOZZI E, PARMAR M, et al. Dopamine Neuron Diversity: Recent Advances and Current Challenges in Human Stem Cell Models and Single Cell Sequencing. Cells. 2021;10(6):1366. [63] NILSSON F, STORM P, SOZZI E, et al. Single-Cell Profiling of Coding and Noncoding Genes in Human Dopamine Neuron Differentiation. Cells. 2021;10(1):137. [64] ÁSGRÍMSDÓTTIR ES, ARENAS E. Midbrain Dopaminergic Neuron Development at the Single Cell Level: In vivo and in Stem Cells. Front Cell Dev Biol. 2020;8:463. [65] BARKER RA, TRANSEURO CONSORTIUM. Designing stem-cell-based dopamine cell replacement trials for Parkinson’s disease. Nat Med. 2019; 25(7):1045-1053. [66] SONNTAG KC, SONG B, LEE N, et al. Pluripotent stem cell-based therapy for Parkinson’s disease: Current status and future prospects. Prog Neurobiol. 2018;168:1-20. [67] ZHANG X, LIU L. Applications of single cell RNA sequencing to research of stem cells. World J Stem Cells. 2019;11(10):722-728. [68] LA MANNO G, GYLLBORG D, CODELUPPI S, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016; 167(2):566-580.e19. [69] HILLER BM, MARMION DJ, THOMPSON CA, et al. Optimizing maturity and dose of iPSC-derived dopamine progenitor cell therapy for Parkinson’s disease. NPJ Regen Med. 2022;7(1):24. [70] CRAIG DW, HUTCHINS E, VIOLICH I, et al. RNA sequencing of whole blood reveals early alterations in immune cells and gene expression in Parkinson’s disease. Nat Aging. 2021;1(8):734-747. [71] AHMED S, KOMEILI M, PARK J. Predictive modelling of Parkinson’s disease progression based on RNA-Sequence with densely connected deep recurrent neural networks. Sci Rep. 2022;12(1):21469. |
| [1] | Yu Shuai, Liu Jiawei, Zhu Bin, Pan Tan, Li Xinglong, Sun Guangfeng, Yu Haiyang, Ding Ya, Wang Hongliang. Hot issues and application prospects of small molecule drugs in treatment of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1913-1922. |
| [2] | Yu Jingbang, Wu Yayun. Regulatory effect of non-coding RNA in pulmonary fibrosis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1659-1666. |
| [3] | Wang Qiuyue, Jin Pan, Pu Rui . Exercise intervention and the role of pyroptosis in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1667-1675. |
| [4] | Yuan Weibo, Liu Chan, Yu Limei. Potential application of liver organoids in liver disease models and transplantation therapy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1684-1692. |
| [5] | Peng Hongcheng, Peng Guoxuan, Lei Anyi, Lin Yuan, Sun Hong, Ning Xu, Shang Xianwen, Deng Jin, Huang Mingzhi . Role and mechanism of platelet-derived growth factor BB in repair of growth plate injury [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1497-1503. |
| [6] | Chen Yilin, Jiang Xiaobo, Qu Honglin, Liu Ruilian. General pattern of GSK3/Nrf2-regulated biological rhythms in organismal aging [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1257-1264. |
| [7] | Lu Ranran, Zhou Xu, Zhang Lijie, Yang Xinling. Dimethyl fumarate alleviates nerve damage in a mouse model of Parkinson’s disease [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 989-994. |
| [8] | Liu Haoyang, Xie Qiang, Shen Mengran, Ren Yansong, Ma Jinhui, Wang Bailiang, Yue Debo, Wang Weiguo . Application, research hotspots, and shortcomings of degradable zinc-based alloys in bone defect repair and reconstruction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(4): 839-845. |
| [9] | Guo Zhao, Zhuang Haoyan, Shi Xuewen. Role of exosomes derived from mesenchymal stem cells in treatment of colorectal cancer [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7872-7879. |
| [10] | Huang Haina, Yu Yanrong, Bi Jian, Huang Miao, Peng Weijie. Epigenetic characteristics of hepatogenic differentiation of mesenchymal stem cells in three-dimensional culture [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7848-7855. |
| [11] | Liu Lu, Zhong Chang, Yu Xin, Ren Chenyuan, Gong Yangyang, Zhou Ping, Wang Yingbin. Academic progress and clinical application of in vitro synthetic microenvironment to promote maturation of human pluripotent stem cell-derived cardiomyocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7856-7862. |
| [12] | Ji Long, Chen Ziyang, , Jin Pan, Kong Xiangkui, Pu Rui, . Lipophagy, exercise intervention and prevention and treatment of nonalcoholic fatty liver disease [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7611-7619. |
| [13] | Yan Jing, Qin Qiujun, Li Fen, Zhou Jun, Ding Yuanyuan, Jin Chunlin. A systematic review of osteoporosis-specific quality-of-life scales [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(35): 7649-7655. |
| [14] | Su Yongkun, Sun Hong, Liu Miao, Yang Hua, Li Qingsong. Development of novel antioxidants and antioxidant combination carried by nano-hydrogel systems in treatment of intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(34): 7376-7384. |
| [15] | Yi Xiaoding, Zhang Di, Guo Hong, Qing Liang, Zhao Tianyu. Decellularized tendon scaffold: a biomedical material for tendon injury repair [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(34): 7385-7392. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||