Chinese Journal of Tissue Engineering Research ›› 2024, Vol. 28 ›› Issue (30): 4909-4914.doi: 10.12307/2024.619
Previous Articles Next Articles
Identification of ferroptosis signature genes in osteoarthritis based on WGCNA and machine learning and experimental validation
Xu Wenfei1, Ming Chunyu2, Duan Kan1, Yuan Changshen1, Guo Jinrong1, Hu Qi1, Zeng Chao1, Mei Qijie1
- 1Orthopedic Department of the Limbs, First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China; 2Department of Geriatrics, Ruikang Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China
-
Received:2023-06-25Accepted:2023-07-24Online:2024-10-28Published:2023-12-28 -
Contact:Mei Qijie, Associate chief physician, Orthopedic Department of the Limbs, First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China -
About author:Xu Wenfei, Master, Orthopedic Department of the Limbs, First Affiliated Hospital of Guangxi University of Chinese Medicine, Nanning 530023, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, No. 82160912 (to DK); National Natural Science Foundation of China, No. 82060875 (to YCS); 2023 Youth Science Fund Project of First Affiliated Hospital of Guangxi University of Chinese Medicine, No. [2023]29 (to XWF)
CLC Number:
Cite this article
Xu Wenfei, Ming Chunyu, Duan Kan, Yuan Changshen, Guo Jinrong, Hu Qi, Zeng Chao, Mei Qijie. Identification of ferroptosis signature genes in osteoarthritis based on WGCNA and machine learning and experimental validation[J]. Chinese Journal of Tissue Engineering Research, 2024, 28(30): 4909-4914.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
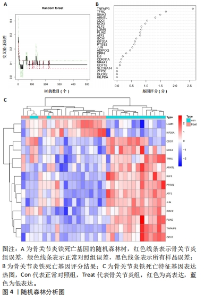
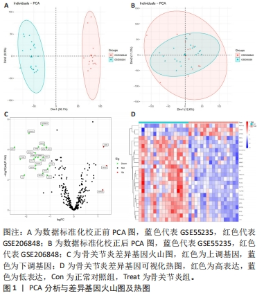
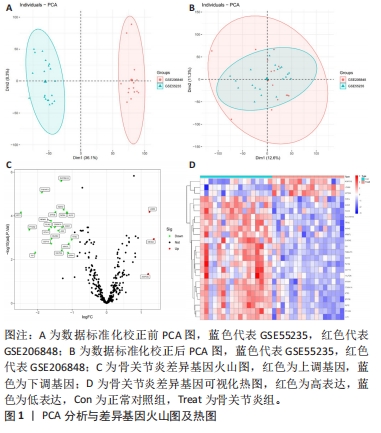
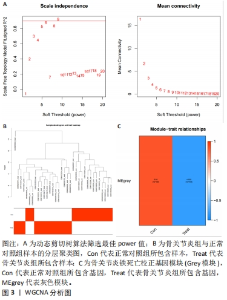
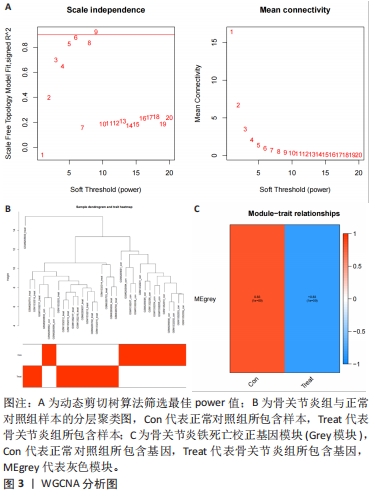
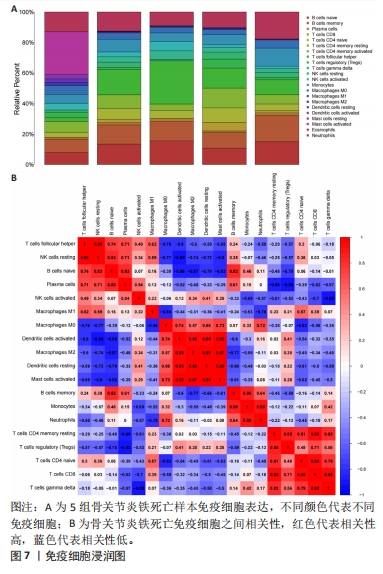
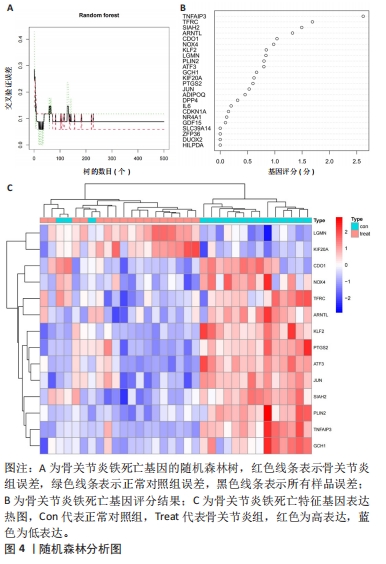
运用R语言对校正基因进行WGCNA分析,通过计算样本间相关性,绘制样本分层聚类图(图3B),同时使用动态剪切树的算法筛选出最佳power值为6(图3A),从而获得一种基因模块(Grey模块),包含有12种特征基因,分别为TNFAIP3、ATF3、LGMN、KLF2、EGR1、PTGS2、CDKN1A、JUN、PLIN2、ZFP36、GCH1、NR4A1(图3C)。 2.4 机器学习 2.4.1 随机森林分析 运用R语言对骨关节炎铁死亡基因进行随机森林分析,以重要性得分大于0.5共获得14种基因,包括TNFAIP3、TFRC、SIAH2、ARNTL、CDO1、NOX4、KLF2、LGMN、PLIN2、ATF3、GCH1、KIF20A、PTGS2、JUN(图4A、B)。同时运用R语言“heatmap”包对获得的基因表达绘制热图,发现TNFAIP3、TFRC、SIAH2、ARNTL、CDO1、NOX4、KLF2、PLIN2、ATF3、GCH1、PTGS2、JUN在正常对照组中呈高表达,而LGMN、KIF20A在骨关节炎组中呈高表达(图4C)。"
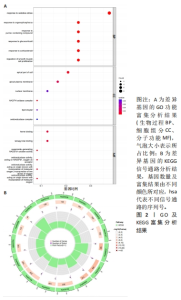
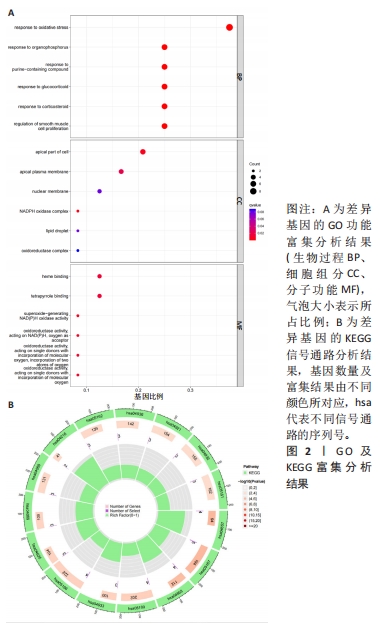
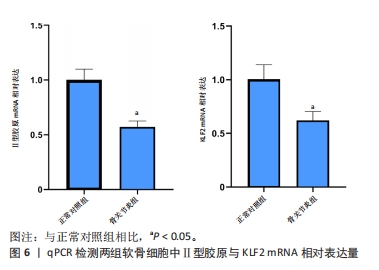
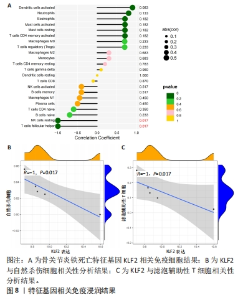
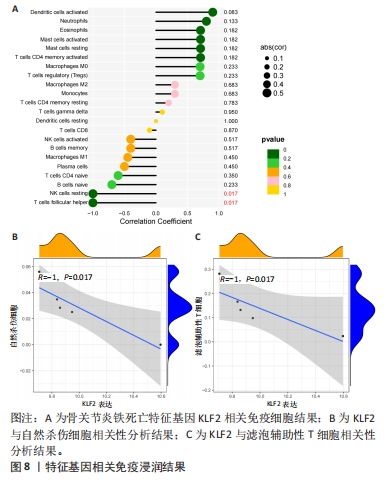
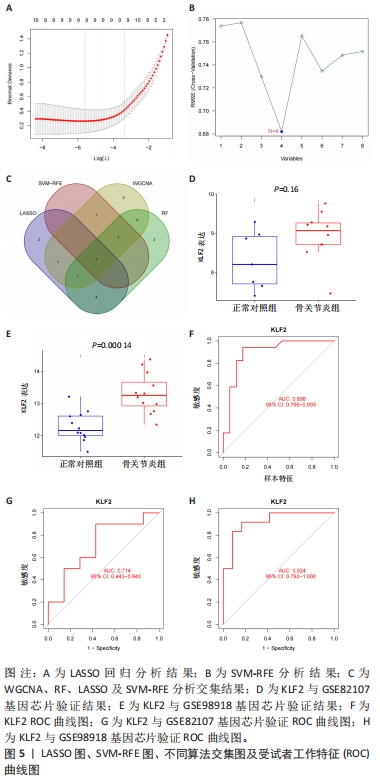
2.4.2 LASSO及SVM-RFE分析 运用R语言对骨关节炎铁死亡基因进行LASSO及SVM-RFE分析。LASSO回归分析筛选9个特征基因(SLC39A14、SIAH2、LGMN、NOX4、KLF2、NR4A1、CDO1、ARNTL、ADIPOQ)(图5A);SVM-RFE分析筛选4个特征基因(ATF3、ZFP36、TNFAIP3、KLF2)(图5B)。 采用R语言“veen”包对WGCNA分析及机器学习(RF、LASSO、SVM-RFE)交集,获得骨关节炎铁死亡特征基因KLF2(图5C);将KLF2与GSE82107、GSE98918基因芯片数据进行验证,发现在滑膜组织中骨关节炎组KLF2的基因表达稍高于正常对照组(P=0.16),但差异无显著性意义(图5D);在半月板组织中骨关节炎组KLF2的基因表达高于正常对照组(P=0.000 14),差异有显著性意义(图5E)。 运用ROC曲线及验证组ROC曲线能更好地判断KLF2作为疾病特征基因的准确性,研究结果显示,ROC:曲线下面积AUC=0.898(图5F),验证组ROC:曲线下面积AUC(GSE82107)=0.714(图5G),曲线下面积AUC(GSE98918)=0.924(图5H),两者均大于0.5,说明KLF2作为疾病特征基因的准确性较高。"
| [1] TANG X, WANG S, ZHAN S, et al. The Prevalence of Symptomatic Knee Osteoarthritis in China: Results From the China Health and Retirement Longitu dinal Study. Arthritis Rheumatol. 2016;68(3):648-653. [2] ZHANG Y, YANG J, YU T, et al. Epidemiological Characteristics of Osteoarthritis in Yichang City-Hubei Province, China, 2017-2018. China CDC Wkly. 2021;3(16): 335-339. [3] CHAILIER E, DEROYER C, CIREGIA F, et al.Chondrocyte dedifferentiation and osteoarthritis(OA). Bioc hem Pharmacol. 2019;165:49-65. [4] JIANG X, STOCKWELL BR, CONRAD M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22(4):266-282. [5] 李哲,袁长深,官岩兵,等.骨关节炎中铁死亡的生物信息学分析与实验验证[J].中国组织工程研究,2023,27(17):2637-2643. [6] CAMACHO DM, COLLINS KM, POWERS RK,et al. Next-Generation Machine Learning for Biological Networks. Cell. 2018;173(7):1581-1592. [7] LIU W, LI L, YE H,et al. Weighted gene co-expression network analysis in biomedicine research. Sheng Wu Gong Cheng Xue Bao. 2017;33(11):1791-1801. [8] WANG F, SU Q, LI C. Identification of cuproptosis-related asthma diagnostic genes by WGCNA analysis and machine learning. J Asthma. 2023;14:1-12. [9] CHEN L, ZHANG YH, WANG S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017; 12(9):0184129. [10] KANEHISA M, FURUMICHI M, TANABE M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(1):353-361. [11] LANGFELDER P, HORVATH S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [12] CHEN X, ISHWARAN H. Random forests for genomic data analysis. Genomics. 2012;99(6):323-329. [13] LI Z, SILLANPAA MJ. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor Appl Genet. 2012; 125(3):419-435. [14] GAO J, KWAN PW, SHI D. Sparse kernel learning with LASSO and Bayesian inference algorithm. Neural Netw. 2010;23(2):257-264. [15] LIVAK KJ, SCHMITTGEN TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25(4):402-408. [16] CHEN D, SHEN J, ZHAO W, et al. Osteoarthritis: toward a comprehensive understanding of pathological mechanism. Bone Res. 2017;5:16044. [17] SACITHARAN PK. Ageing and Osteoarthritis. Subcell Biochem. 2019;91:123-159. [18] HAWKER GA, KING LK. The Burden of Osteoarthritis in Older Adults. Clin Geriatr Med. 2022;38(2):181-192. [19] OO WM. Prospects of Disease-Modifying Osteoarthritis Drugs. Clin Geriatr Med. 2022;38(2):397-432. [20] YAO X, SUN K, YU S, et al. Chondrocyte ferroptosis contribute to the progressi on of osteoarthritis. J Orthop Translat. 2020;27:33-43. [21] STOCKWELL BR, FRIEDMANN ANGELI JP, BBYIR H, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017; 171(2):273-285. [22] JING X, LIN J, DU T, et al. Iron Overload Is Associated With Accelerated Progression of Osteoarthritis: The Role of DMT1 Mediated Iron Homeostasis. Front Cell Dev Biol. 2021;8:594509. [23] JING X, DU T, LI T, et al.The detrimental effect of iron on OA chondrocytes: Importance of pro-inflammatory cytokines induced iron influx and oxidative stress. J Cell Mol Med. 2021;25(12):5671-5680. [24] ZAHAN OM, SERBAN O, GHERMAN C, et al. The evaluation of oxidative stress in osteoarthritis. Med Pharm Rep. 2020;93(1):12-22. [25] BONJOUR JP. Calcium and phosphate: a duet of ions playing for bone health. J Am Coll Nutr. 2011;30(5):438-448. [26] NA HS, PARK JS, CHO KH, et al. Interleukin-1-Interleukin-17 Signaling Axis Induces Cartilage Destruction and Promotes Experimental Osteoarthritis. Front ImMunol. 2020;11:730. [27] CAI P, JIANG T, LI B, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (OA) based on microarray profiles of human joint fibroblast-like synoviocytes. Cell Biochem Funct. 2019;37(1):31-41. [28] WITTNER J, SCHUH W. Krüppel-like Factor 2 (KLF2) in Immune Cell Migration. Vaccines (Basel). 2021;9(10):1171. [29] KAWATA M, TERAMURA T, ORDOUKHANIAN P, et al. Krüppel-like factor-4 and Krüppel-like factor-2 are important regulators of joint tissue cells and protect against tissue destruction and inflammation in osteoarthritis. Ann Rheum Dis. 2022;9:221867. [30] GAO X, JIANG S, DU Z, et al. KLF2 Protects against Osteoarthritis by Repressing Oxidative Response through Activation of Nrf2/ARE Signaling In Vitro and In Vivo. Oxid Med Cell Longev. 2019;19:8564681 [31] YUAN Y, TAN H, DAI P. Krüppel-Like Factor 2 Regulates Degradation of Type II Collagen by Suppressing the Expression of Matrix Metalloproteinase (MMP)-13. Cell Physiol Biochem. 2017;42(6):2159-2168. [32] LAHA D, DEB M, DAS H. KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy. Autophagy. 2019;15(12):2063-2075. [33] HOU Z, WANG Z, TAO Y, et al. KLF2 regulates osteoblast differentiation by targeting of Runx2. Lab Invest. 2019;99(2):271-280. [34] RAGIPOGLU D, DUDECK A, HAFFNER-LUNTZER M, et al. The Role of Mast Cells in Bone Metabolism and Bone Disorders. Front Immunol. 2020;11:163. [35] GERMIC N, FRANGEZ Z, YOUSEFI S, et al. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 2019;26(4):715-727. [36] CAI W, LI H, ZHANG Y, et al. Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. Peer J. 2020,8:8390. [37] FERNANDES JC, MARTEL-PELLETIER J, PELLETIER JP. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237-246. |
| [1] | Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. The construction and validation of a prediction model based on multiple machine learning algorithms and the immunomodulatory analysis of rheumatoid arthritis from the perspective of mitophagy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-15. |
| [2] | Ma Chi, Wang Ning, Chen Yong, Wei Zhihan, Liu Fengji, Piao Chengzhe. Application of 3D-printing patient-specific instruments combined with customized locking plate in opening wedge high tibial osteotomy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1863-1869. |
| [3] | Yu Shuai, Liu Jiawei, Zhu Bin, Pan Tan, Li Xinglong, Sun Guangfeng, Yu Haiyang, Ding Ya, Wang Hongliang. Hot issues and application prospects of small molecule drugs in treatment of osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1913-1922. |
| [4] | Zhao Jiyu, Wang Shaowei. Forkhead box transcription factor O1 signaling pathway in bone metabolism [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1923-1930. |
| [5] | Sun Yundi, Cheng Lulu, Wan Haili, Chang Ying, Xiong Wenjuan, Xia Yuan. Effect of neuromuscular exercise for knee osteoarthritis pain and function: a meta-analysis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1945-1952. |
| [6] | Deng Keqi, Li Guangdi, Goswami Ashutosh, Liu Xingyu, He Xiaoyong. Screening and validation of Hub genes for iron overload in osteoarthritis based on bioinformatics [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1972-1980. |
| [7] | Yin Lu, Jiang Chuanfeng, Chen Junjie, Yi Ming, Wang Zihe, Shi Houyin, Wang Guoyou, Shen Huarui. Effect of Complanatoside A on the apoptosis of articular chondrocytes [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1541-1547. |
| [8] | Wang Peiguang, Zhang Xiaowen, Mai Meisi, Li Luqian, Huang Hao. Generalized equation estimation of the therapeutic effect of floating needle therapy combined with acupoint embedding on different stages of human knee osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1565-1571. |
| [9] | Wang Qiuyue, Jin Pan, Pu Rui . Exercise intervention and the role of pyroptosis in osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1667-1675. |
| [10] | Chen Yueping, Chen Feng, Peng Qinglin, Chen Huiyi, Dong Panfeng . Based on UHPLC-QE-MS, network pharmacology, and molecular dynamics simulation to explore the mechanism of Panax notoginseng in treating osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(8): 1751-1760. |
| [11] | Yang Zhihang, Sun Zuyan, Huang Wenliang, Wan Yu, Chen Shida, Deng Jiang. Nerve growth factor promotes chondrogenic differentiation and inhibits hypertrophic differentiation of rabbit bone marrow mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1336-1342. |
| [12] | Zhao Nannan, Li Yanjie, Qin Hewei, Zhu Bochao, Ding Huimin, Xu Zhenhua. Changes in ferroptosis in hippocampal neurons of vascular dementia model rats treated with Tongmai Kaiqiao Pill [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1401-1407. |
| [13] | Zhang Mingyang, Yang Xinling. Verbascoside inhibits Erastin-induced ferroptosis of dopaminergic nerve cell line MN9D cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1408-1413. |
| [14] | Wang Mi, Ma Shujie, Liu Yang, Qi Rui. Identification and validation of characterized gene NFE2L2 for ferroptosis in ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(7): 1466-1474. |
| [15] | Qian Kun, Li Ziqing, Sun Shui . Endoplasmic reticulum stress in the occurrence and development of common degenerative bone diseases [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(6): 1285-1295. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||