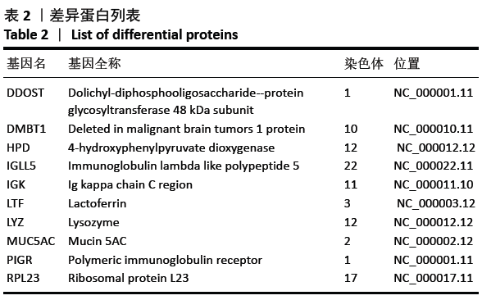
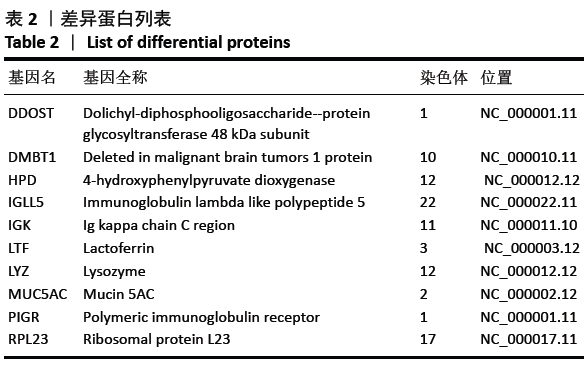
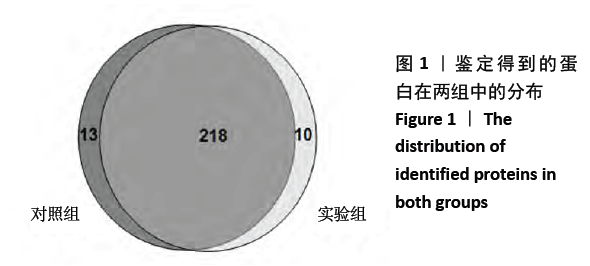
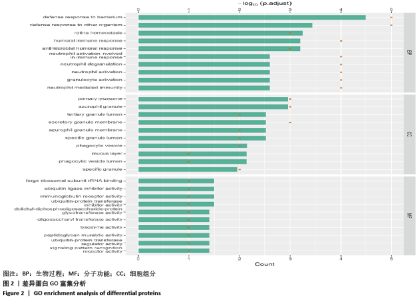
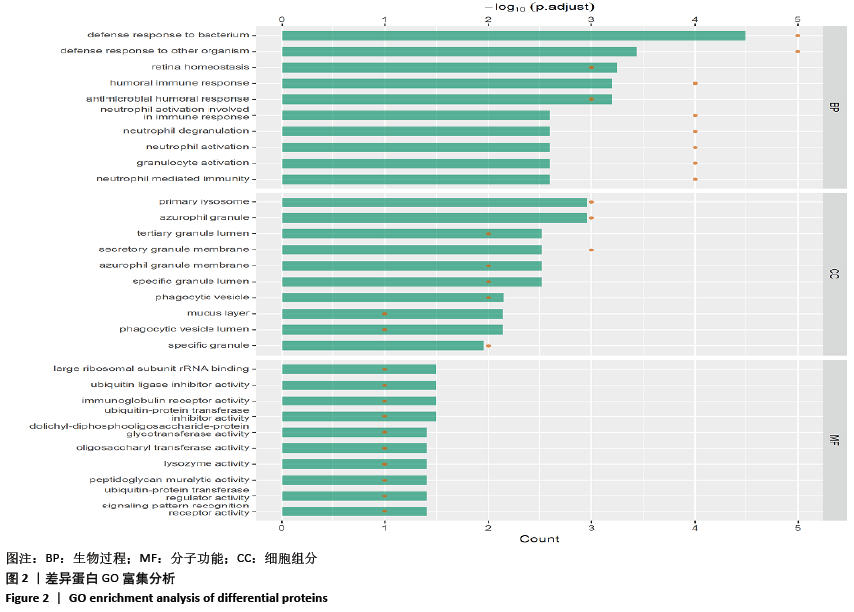
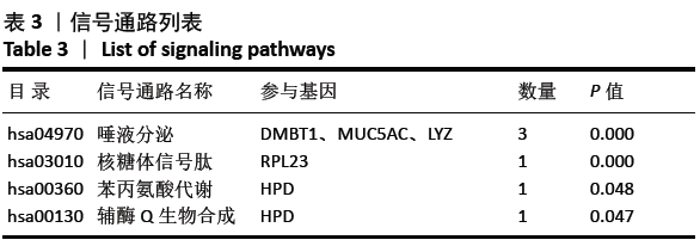
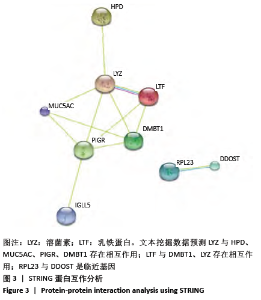
[1] SOWERS MR, ZHENG H, GREENDALE GA, et al. Changes in bone resorption across the menopause transition: effects of reproductive hormones, body size, and ethnicity. J Clin Endocrinol Metab. 2013; 98(7):2854-2863.
[2] DJEBALI S, DAVIS CA, MERKEL A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101-108.
[3] HUYNH NP, ANDERSON BA, GUILAK F, et al. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58(1):116-141.
[4] MA Z, GUO C. Advances on the effects of lncRNA on bone metabolism related signaling pathways in Osteoporosis. Chin J Osteoporos. 2018; 4(4):547-551.
[5] WU QY, LI X, MIAO ZN, et al. Long Non-coding RNAs: A New Code for Osteoporosis. Front Endocrinol. 2018;9:587.
[6] SILVA AM, MOURA SR, TEIXEIRA JH, et al. Long noncoding RNAs: a missing link in osteoporosis. Bone Res. 2019;7:10.
[7] ZHANG HL, DU XY, DONG QR. LncRNA XIXT promotes osteogenic differentiation of bone mesenchymal stem cells and alleviates osteoporosis progression by targeting miRNA-30a-5p. Eur Rev Med Pharmacol Sci. 2019;23(20):8721-8729.
[8] HAN Y, LIU C, LEI M, et al. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J Orthop Surg Res. 2019;14(1):416
[9] LING L, HU HL, LIU KY, et al. Long noncoding RNA MIRG induces osteoclastogenesis and bone resorption in osteoporosis through negative regulation of miR-1897. Eur Rev Med Pharmacol Sci. 2019; 23(23):10195-10203.
[10] CHEN J, XIE L, LI S, et al. Analysis of lncRNA expression and regulation network in postmenopausal osteoporosis patients with Kidney Yin deficiency. Chin J Osteoporos. 2015;21(5):553-559.
[11] LI S, XU H, CHEN J, et al. Study on the expression of lincRNA uc431+ in postmenopausal osteoporosis patients with kidney-yin deficiency syndrome. Chin J Osteoporos. 2016;22(8):966-971.
[12] SETA N, OKAZAKI Y, KUWANA M. Human circulating monocytes can express receptor activator of nuclear factor-kappaB ligand and differentiate into functional osteoclasts without exogenous stimulation. Immunol Cell Biol. 2008;86(5):453-459.
[13] WANG H, SHEN Y. MicroRNA 20a negatively regulates the growth and osteoclastogenesis of THP 1 cells by downregulating PPARγ. Mol Med Rep. 2019;20(5):4271-4276.
[14] MOULINE CC, QUINCEY D, LAUGIER JP, et al. Osteoclastic differentiation of mouse and human monocytes in a plasma clot/biphastic calcium phosphate microparticles composite. European cells and materials. 2010;20:379-392.
[15] SZKLARCZYK D, GABLE AL, LYON D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-613.
[16] OKAZAKI Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):63-73.
[17] 熊坤,邓江,黄文良,等.长链非编码RNA参与调控成骨细胞分化和破骨细胞的生成[J].中国组织工程研究,2020,24(14):2229-2234.
[18] WANG H, ZHAO W, TIAN QJ, et al. Effect of lncRNA AK023948 on rats with postmenopausal osteoporosis via PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(5):2181-2188.
[19] WANG H, LI YK, CUI M, et al. Effect of lncRNA AK125437 on postmenopausal osteoporosis rats via MAPK pathway. Eur Rev Med Pharmacol Sci. 2020;24(5):2173-2180.
[20] FAN P, FENG XY, HU N, et al. Expression of long-chain non-coding RNA NEAT1 in osteoporosis and its effect on differentiation of osteoblasts and osteoclasts. Joural of clinical and experimental medicine. 2019;18 (23):2504-2507.
[21] ZHANG J, HAO X, YIN M, et al. Long non-coding RNA in osteogenesis: A new world to be explored. Bone Joint Res. 2019;8(2):73-80.
[22] VICENTINI C, GALUPPINI F, CORBO V, et al. Current role of non-coding RNAs in the clinical setting. Noncoding RNA Res. 2019;4(3):82-85.
[23] GHAFOURI-FARD S, TAHERI M. Long non-coding RNA signature in gastric cancer. Exp Mol Pathol. 2019;113:104365.
[24] HAN Y, LIU C, LEI M, et al. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J Orthop Surg Res. 2019;14(1):416.
[25] 孙强,赵轶男,王远瑞,等. LncRNA HOTAIR调控miR-130a-3p对骨关节炎软骨细胞增殖和分化的影响[J].中国骨与关节杂志,2019, 8(12):931-937.
[26] CAITLIN AC, ELIZABETH AM, JAMES DM. Production of Human Lactoferrin and Lysozyme in the Milk of Transgenic Dairy Animals: Past, Present, and Future. Transgenic Res. 2015;24(4):605-614.
[27] THOMAS JH, GARY M, ARNOLD JK. Lysozyme synthesis in osteoclasts. J Bone Miner Res. 1990;12(5):1217-1222.
[28] DAGENG H , YANGYANG W, JING L, et al. Proteomic profiling analysis of postmenopausal osteoporosis and osteopenia identifies potential proteins associated with low bone mineral density. PeerJ. 2020;8: e9009.
[29] CORNISH J, CALLON KE, NAOT D, et al. Lactoferrin is a potent regulator of bone cell activity and increases bone formation in vivo. Endocrinology. 2004;145(9):4366-4374.
[30] LIU M, FAN F, SHI P, et al. Lactoferrin promotes MC3T3-E1 osteoblast cells proliferation via MAPK signaling pathways. Int J Biol Macromol. 2018;107(Pt A):137-143
[31] FAN F, SHI P, LIU M, et al. Lactoferrin preserves bone homeostasis by regulating the RANKL/RANK/OPG pathway of osteoimmunology. Food Funct. 2018;9(5):2653-2660.
[32] CHEN XW, LI YH , ZHANG MJ, et al. Lactoferrin ameliorates aging-suppressed osteogenesis via IGF1 signaling.J Mol Endocrinol. 2019; 63(1):62-75.
[33] XU Y, AN JJ, TABYS D, et al. Effect of Lactoferrin on the Expression Profiles of Long Non-coding RNA during Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Int J Mol Sci. 2019;20(19) pii: E4834.
[34] LI Y, ZHANG W2, REN F, et al. Activation of TGF-β Canonical and Noncanonical Signaling in Bovine Lactoferrin-Induced Osteogenic Activity of C3H10T1/2 Mesenchymal Stem Cells. Int J Mol Sci. 2019; 20(12) pii: E2880.
[35] HANSTOCK HG, EDWARDS JP, WALSH NP. Tear Lactoferrin and Lysozyme as Clinically relevant biomarkers of mucosal immune competence. Front Immunol. 2019;10:1178.
[36] ANDRÉ GO, POLITANO WR , MIRZA S,et al. Combined effects of lactoferrin and lysozyme on streptococcus pneumoniae killing. Microb pathog . 2015;89:7-17.
[37] VAN BERKEL PH, GEERTS ME, VAN VEEN HA, et al. N-terminal stretch Arg2, Arg3, Arg4 and Arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem J. 1997;328(Pt 1):145-151
[38] XIE L, FAN C, LI S, et al. Effects of liuwei dihuang pill on difference-expressed genes of kidney Yin deficiency syndrome in postmenopausal osteoporosis. Chin J Osteoporos. 2015;21(8):971-976+985.
[39] SHEN Z, WANG W. Reference standard for syndrome differentiation of deficiency syndrome in traditional Chinese medicine. Chin J Integr Med.1986;6(10):598.
[40] CHANG Z,FEN C,SHI X,et al.Research progress of immunity and osteoporosis.Chin J Osteoporos. 2015;21(4):508-513.
[41] LI S, FENG E, ZHANG Y, et al. Study of the gene expression profile in the bone tissue with kidney yin deficiency syndromes in primary osteoporosis. Chin J Osteoporos. 2013;19(12):1215-1218+1253.
|