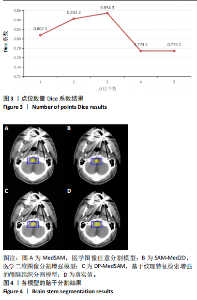
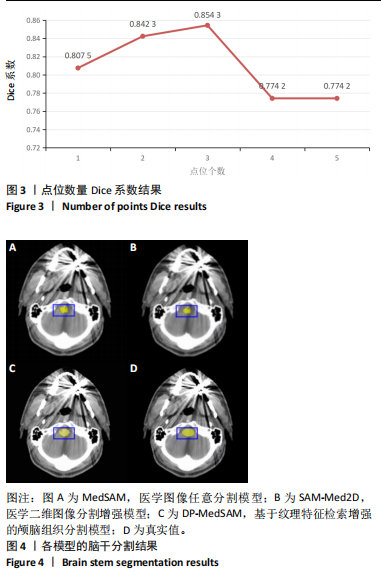
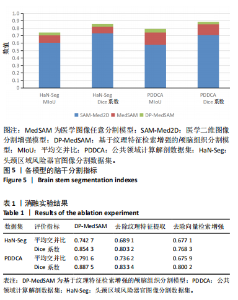
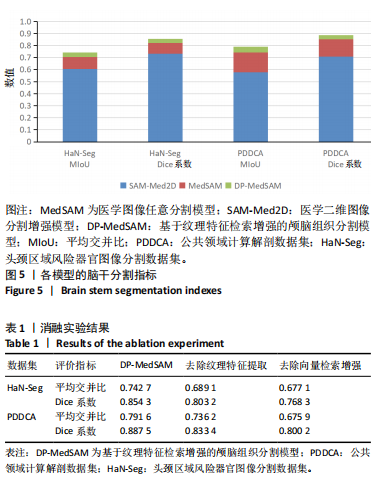
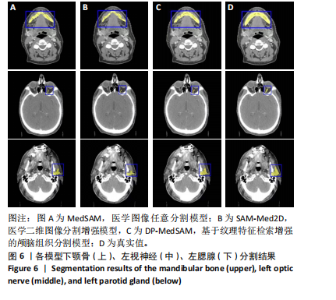
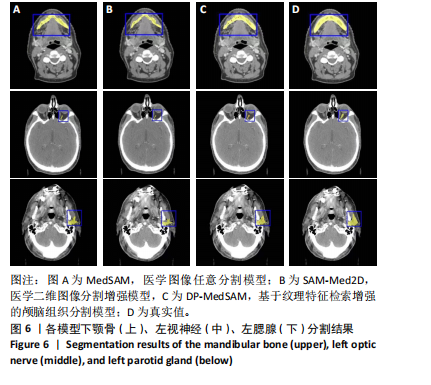
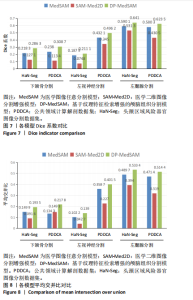
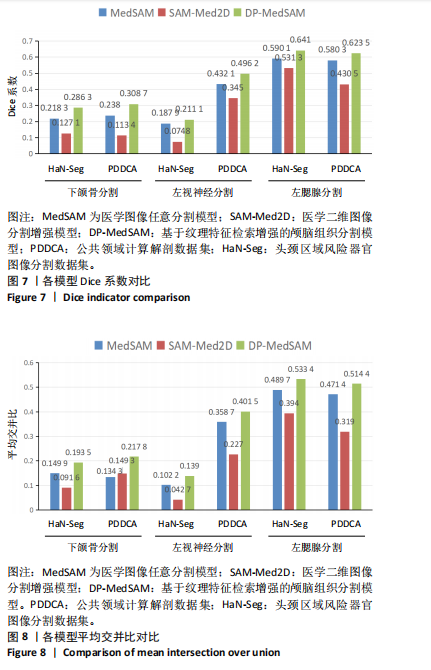
[1] HUANG YH, YANG X, LIU L, et al. Segment Anything Model for Medical Images? Med Image Anal. 2024;92:103061-103061.
[2] ALI M, WU T, HU H, et al. A review of the Segment Anything Model (SAM) for medical image analysis: Accomplishments and perspectives. Comput Med Imaging Graph. 2024;119:102473.
[3] GUO A, FEI G, PASUPULETI H, et al. ClickSAM: Fine-tuning Segment Anything Model using click prompts for ultrasound image segmentation. Proc SPIE Int Soc Opt Eng. 2024;12932:1293212.
[4] 阚明阳. 基于深度学习在医学图像的分类与诊断综述[J]. 新一代信息技术,2021,4(20):17-22.
[5] GU YB, WANG QY, TANG H, et al. LeSAM: Adapt Segment Anything Model for Medical Lesion Segmentation. IEEE JBHI. 2024;28(10)1-11.
[6] KOYAMA Y, NAKASHIMA K, ORIHARA S, et al. Inter- and intra-observer variability of qualitative visual breast-composition assessment in mammography among Japanese physicians: a first multi-institutional observer performance study in Japan. Breast Cancer. 2024;31(4):671-683.
[7] MA J, HE YT, LI FF, et al. Segment Anything in Medical Images. Nat Commun. 2024;15(1):1-9.
[8] SUN J, CHEN K, HE Z, et al. Medical image analysis using improved SAM-Med2D: segmentation and classification perspectives. BMC Med Imaging. 2024;24(1):241.
[9] 张亚楠,张伟方.院前序贯评估路径表在颅脑损伤患者救治中的应用价值[J].中国医药导报,2023,20(10):165-169.
[10] KANG J, BENJAMIN DI, KIM S, et al. Depletion of SAM leading to loss of heterochromatin drives muscle stem cell ageing. Nat Metab. 2024;6(1):153-168.
[11] LV Y, CHANG J, ZHANG W, et al. Improving Microbial Cell Factory Performance by Engineering SAM Availability. J Agric Food Chem. 2024;72(8):3846-3871.
[12] CHEEK LE, ZHU W. Structural features and substrate engagement in peptide-modifying radical SAM enzymes. Arch Biochem Biophys. 2024;756:110012.
[13] 尹红云,张丽娜,王佳明,等.基于注意力机制和深度学习的颅脑外伤患者CT图像分割[J].生命科学仪器,2024,22(1):20-22.
[14] 杜冉,李文娟,冯方,等. 血小板/淋巴细胞比值对重度创伤性颅脑损伤患者30d内病死风险的预测价值[J]. 郑州大学学报(医学版),2024,59(3):373-376.
[15] 方诚. 基于机器学习方法建立预测模型用于创伤性颅脑损伤领域的研究[D]. 合肥:安徽医科大学,2023.
[16] 李哲. 基于深度学习的脑微出血检测方法研究[D]. 沈阳:沈阳理工大学,2023.
[17] 王聪. 基于乘员伤害分析的颅脑精细建模与损伤机理研究[D]. 长春:吉林大学,2015.
[18] PODOBNIK G, STROJAN P, PETERLIN P, et al. HaN-Seg: the Head and Neck Organ-at-risk CT and MR Segmentation Challenge. Medical Physics. 2023;50(3):1917-1927.
[19] RAUDASCHL PF, ZAFFINO P, SHARP GC, et al. Evaluation of Segmentation Methods on Head and Neck CT: Auto-segmentation Challenge 2015. Med Phys. 2017;44(5):2020-2036.
[20] WEI XP, YU XC, LIU B, et al. Convolutional Neural Networks and Local Binary Patterns for Hyperspectral Image Classification. Eur J Rem Sens. 2019;52(1):448-462.
[21] TANG D, JIANG X, WANG K, et al. Toward identity preserving in face sketch-photo synthesis using a hybrid CNN-Mamba framework. Sci Rep. 2024;14(1):22495.
[22] GUO ZH, ZHANG L, ZHANG D. Rotation invariant texture classification using LBP variance (LBPV) with global matching. Pattern Recogn. 2010;43(3): 706-719.
[23] 徐先传,张琦.基于LBP算子与EMD距离的医学图像检索[J].微计算机信息,2009,25(9):275-276+295.
[24] 朱碧云,陈卉.医学图像纹理分析的方法及应用[J].中国医学装备, 2013,10(8):77-81.
[25] TAIPALUS T. Vector Database Management Systems: Fundamental Concepts, Use-Cases, and Current Challenges. Cogn Syst Res. 2024;85: 101216.
[26] ESWARAIAH P, SYED H. Deep learning-based information retrieval with normalized dominant feature subset and weighted vector model. PeerJ Comput Sci. 2024;10:e1805.
[27] 张浩.基于向量数据库的智能媒资搜索研究[J].电声技术,2022, 46(1):22-24+28.
[28] RONNEBERGER O, FISCHER P, BROX T. U-Net: Convolutional Networks for Biomedical Image Segmentation. MICCAI. 2015; 9351:234-241.
[29] SU Q, HE W, WEI X, et al. Multi-scale full spike pattern for semantic segmentation. Neural Netw. 2024;176:106330.
[30] BUEHLER MJ. Generative Retrieval-Augmented Ontologic Graph and Multiagent Strategies for Interpretive Large Language Model-Based Materials Design. ACS Eng Au. 2024;4(2):241-277.
[31] LIU D, LI H, ZHAO Z, et al. Text-guided Image Restoration and Semantic Enhancement for Text-to-Image Person Retrieval. Neural Netw. 2024; 184:107028.
[32] JIAO R, ZHANG Y, DING L, et al. Learning with limited annotations: A survey on deep semi-supervised learning for medical image segmentation. Comput Biol Med. 2024;169:107840.
[33] RIAAN Z, EFSTRATIOS G, MATTHAN C, et al. Domain- and Task-Specific Transfer Learning for Medical Segmentation Tasks. Comput Methods Programs Biomed. 2022;214:106539.
[34] WEN HL, SONG Y, YAN W, et al. Multi-level similarity learning for image-text retrieval. Inform Proc Manag. 2021;58(1):102432.
[35] FU Z, FU S, HUANG Y, et al. Application of large language model combined with retrieval enhanced generation technology in digestive endoscopic nursing. Front Med (Lausanne). 2024;11:1500258.
|