Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (26): 5595-5607.doi: 10.12307/2025.745
Previous Articles Next Articles
Rheumatoid arthritis from the perspective of mitophagy: interaction analysis based on multiple machine learning algorithms
Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong
- Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China
-
Received:2024-08-01Accepted:2024-09-15Online:2025-09-18Published:2025-02-25 -
Contact:Chen Yueping, PhD, Chief physician, Doctoral supervisor, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China -
About author:Li Jiagen, Master candidate, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning 530011, Guangxi Zhuang Autonomous Region, China -
Supported by:National Natural Science Foundation of China, No. 82360937 (to CYP); Natural Science Foundation of Guangxi Zhuang Autonomous Region, No. 2023JJA140318 (to CYP); Guangxi Integrated Bone and Joint Degenerative Disease Multidisciplinary Innovation Team Project, No. GZKJ2310 (to CYP [project participant]); Innovation Project of Guangxi Graduate Education of Guangxi University of Chinese Medicine, No. YCSY2023040 (to LJG)
CLC Number:
Cite this article
Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. Rheumatoid arthritis from the perspective of mitophagy: interaction analysis based on multiple machine learning algorithms[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(26): 5595-5607.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
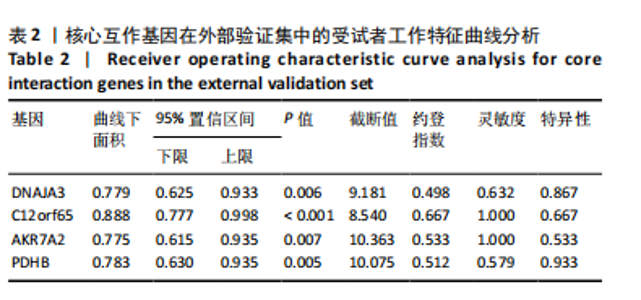
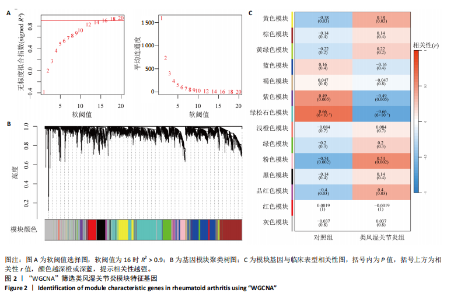
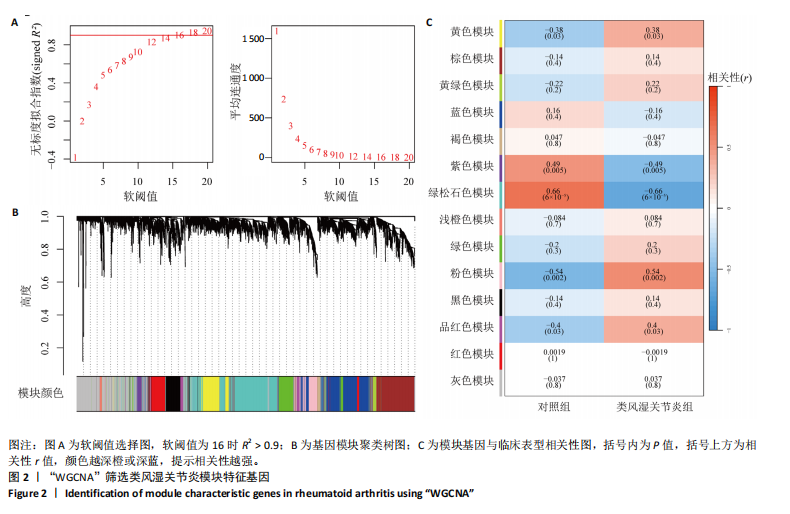
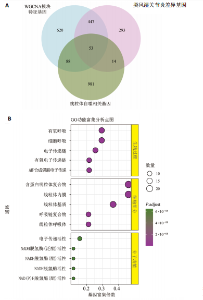
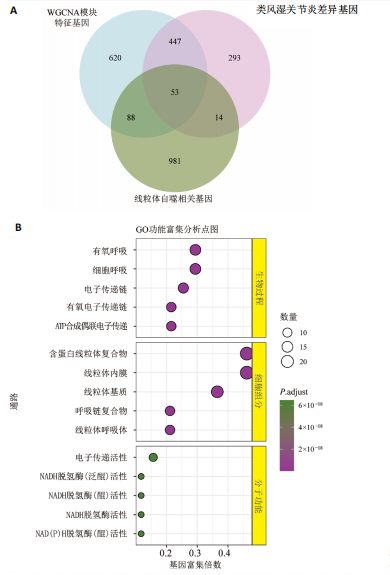
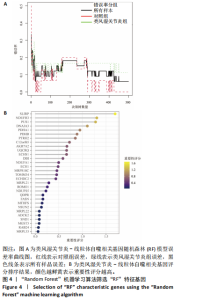
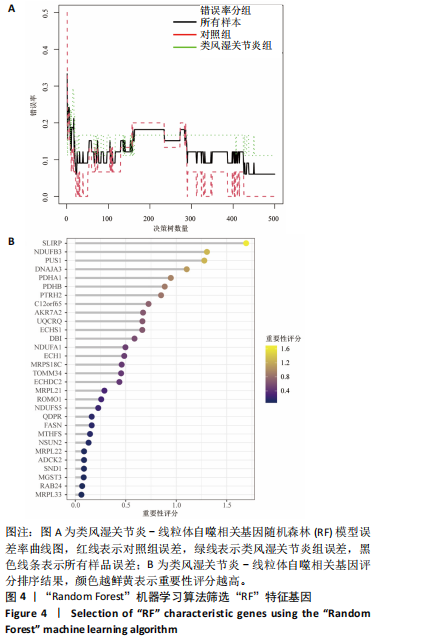
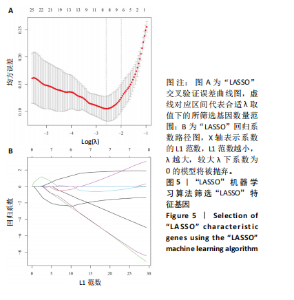
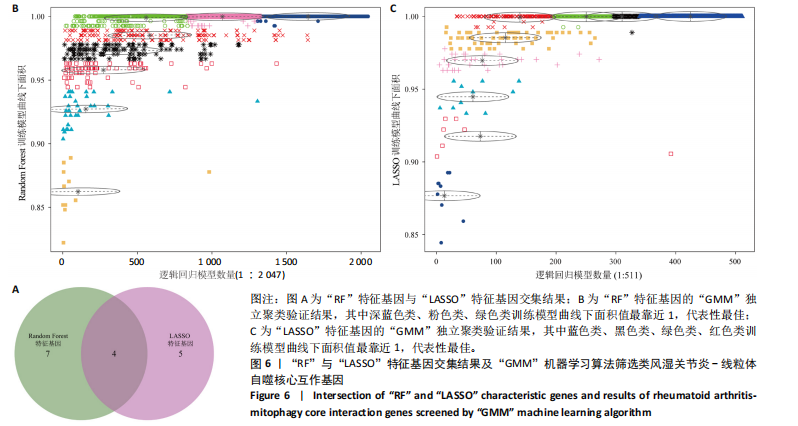
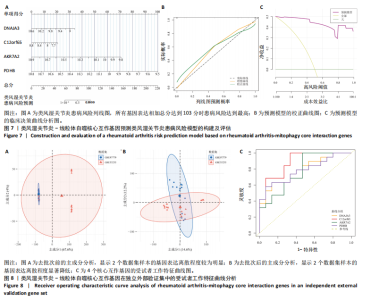
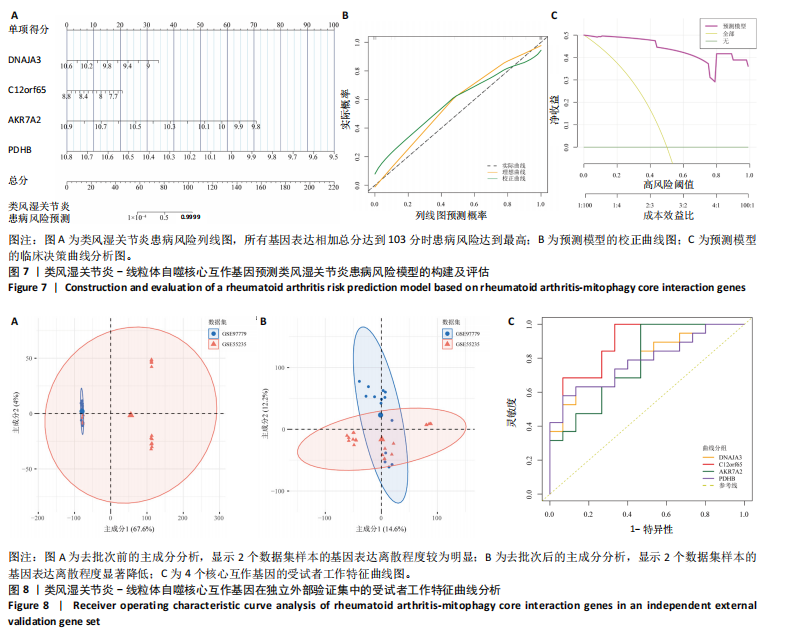
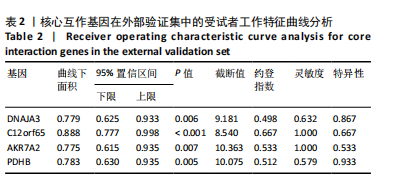
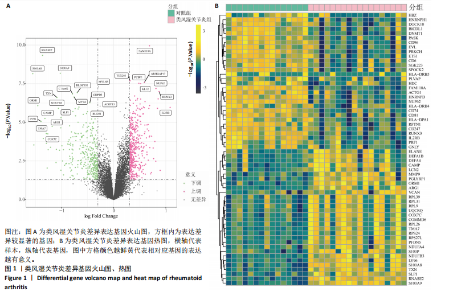
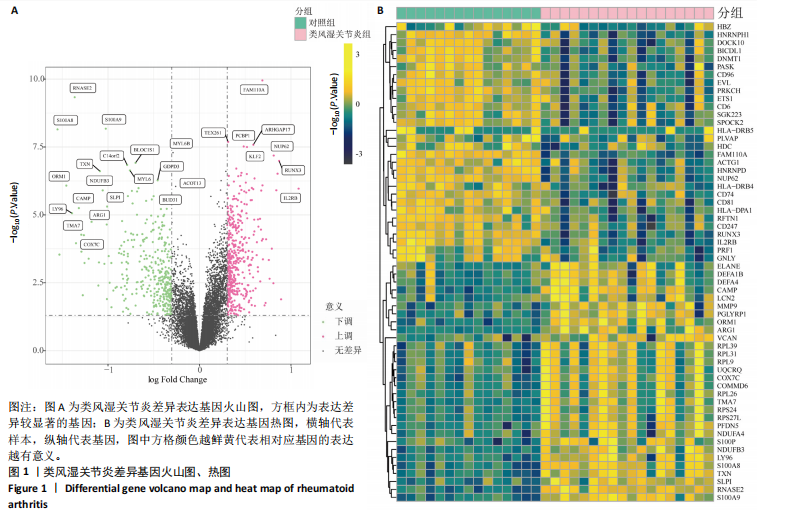
2.1 类风湿关节炎差异表达基因筛选结果 通过R函数包“limma”和“affy”分析类风湿关节炎组和正常组的基因转录组数据,并根据平均绝对折叠变化法获得的log FC Cut Off及-log10 (P.Value) < 0.05,筛选差异基因总计807个,其中上调基因数量为439个,下调基因数量为368个。将差异基因进行火山图可视化(图1A),并对上调及下调基因中表达量最高的各30位基因进行热图可视化(图1B)。 2.2 “WGCNA”分析结果 “WGCNA”可作为一种初步筛选重要性基因的强大方法,通过对标准化后的矩阵数据进一步处理,发现数据集中无不良样本及基因被删除,然后对所有样本聚类,发现GSM389724与GSM389732两例样本离群,当“cutHeight”=45时,切除离群样本。通过计算不同软阈值的“scale free topology model fit”,继而构建无标度网络(图2A)。随后,集合不同表达水平基因的基因模块被分离出来,通过聚类分析和动态剪切树法识别模块,并合并相似的模块(图2B)。计算模块特征基因与临床表型(正常对照和类风湿关节炎)之间的相关性,生成相关性矩阵和P值矩阵,继而绘制模块与临床表型的相关性热图(图2C) ,结果显示绿松石色模块(r=±0.66,P=6×10-5)、粉色模块(r=±0.54,P=0.002)意义最显著。提取上述模块基因,其中绿松石色模块含基因970个,粉色模块含基因238个,共有1 208个模块特征基因与临床表型最相关。 2.3 类风湿关节炎-线粒体自噬相关基因筛选及“GO”和“KEGG”分析结果 从“MitoCarta 3.0”数据库中下载人类线粒体特征表达基因,数据集包含1 136个基因,以807个类风湿关节炎差异表达基因、1 208个“WGCNA”特征基因和人类线粒体1 136个特征表达基因取交集,获得类风湿关节炎-线粒体自噬相关基因总计53个(图3A)。对上述交集基因进行“GO”和“KEGG”功能富集分析,“GO”富集分析排行前5位的可视化结果显示生物过程主要与有氧呼吸、细胞呼吸、电子传递链、有氧电子传递链、ATP合成偶联电子传递有关,细胞组分主要与含蛋白线粒体复合物、线粒体内膜、线粒体基质、呼吸链复合物、线粒体呼吸体相关,分子功能主要与电子传递活性、NADH脱氢酶(泛醌)活性、NADH脱氢酶(醌)活性、NADH脱氢酶活性、NAD(P)H脱氢酶(醌)活性有关(图3B)。“KEGG”功能富集分析结果包含4条细胞过程通路,富集程度由高到低依次为线粒体自噬、过氧化物酶体代谢、细胞衰老、坏死性凋亡(图3C)。 2.4 “RF”机器学习算法筛选核心诊断基因 采用R函数包“randomForest”的“RF”算法对53个类风湿关节炎-线粒体自噬相关基因进行进一步筛选并对随机森林模型误差率曲线图(图4A)和基因重要性排名图(图4B)可视化,获得30个相关性较高的基因,当重要性评分> 0.6时,获得“RF”算法相关性最佳的11个基因,分别为SLIRP、NDUFB3、PUS1、DNAJA3、PDHA1、PDHB、PTRH2、C12orf65、AKR7A2、UQCRQ、ECHS1。 2.5 “LASSO”机器学习算法筛选核心诊断基因 通过R函数包“tidyverse”“broom”和“glmnet”对53个类风湿关节炎-线粒体自噬相关基因进行“LASSO”回归分析,绘制出“LASSO”回归模型交叉验证误差曲线图(图5A)和“LASSO”回归系数路径图(图5B)。根据交叉验证误差曲线图可知,“LASSO”筛选的特征基因数量在6-9之间,输出基因筛选结果,获得9个相关性最高的基因,包括DNAJA3、C12orf65、AKR7A2、HADH、PDHB、BLOC1S1、COX7C、SLC25A5、NDUFS5。 2.6 “GMM”机器学习算法同级验证核心互作基因 将“RF”算法与“LASSO”算法筛选结果取交集,进行Venn图可视化(图6A),取得4个交集基因分别为DNAJA3、C12orf65、AKR7A2、PDHB。通过R函数包“mclust”和“SimDesign”对“RF”和“LASSO”回归分析筛选出的特征基因分别进行“GMM”独立聚类验证,“RF”和“LASSO”的基因筛选结果分别被归聚为9个类别,验证“RF”结果下创建了2 047个训练模型(图6B);验证“LASSO”结果下创建了511个训练模型(图6C)。输出两侧最佳模型结果,包括DNAJA3、PUS1、AKR7A2、PTRH2、ECHS1、PDHB、C12orf65、SLC25A5、COX7C共9个基因,其中DNAJA3、C12orf65、AKR7A2、PDHB均位于“GMM”算法下最优训练模型基因结果内,因此被筛选作为4个类风湿关节炎-线粒体自噬核心互作基因。 2.7 预测模型构建及验证结果 在二分类逻辑回归分析模式下,对4个类风湿关节炎-线粒体自噬核心互作基因进行诊断列线图模型构建,列出各个基因的分数区间,以用来评估基因表达水平在不同分数下的风险贡献,并通过相加每个基因的分数计算出总分,通过总分评估相对应的风险概率。总分越高,患类风湿关节炎的风险越大(图7A)。同时基于Bootstrap方法,重复1 000次,进行模型校正曲线绘制(图7B),以进行内部验证,结果发现校正曲线与理想曲线贴合度高(均方误差=0.005 15),证明模型的构建精度良好。进一步构建临床决策曲线(图7C),发现预测模型曲线与所有样本患病曲线之间离散度较高,表明模型的临床效用较高。 2.8 外部患者样本的工作特征曲线验证 将GSE97779与GSE55235数据集去批次合并获得外部患者样本的独立验证集,通过对比去批次前后的主成分分析(图8A,B),发现去批次效果良好。在独立外部验证集基因表达数据中进行核心互作基因的受试者工作特征曲线分析(表2)并进行可视化(图8C),结果发现所有核心互作基因的曲线下面积均大于0.7 (P < 0.01),表明诊断效能良好,证明4个类风湿关节炎-线粒体自噬核心互作基因在实际应用中对类风湿关节炎的预测具有良好的稳定性与准确性。"
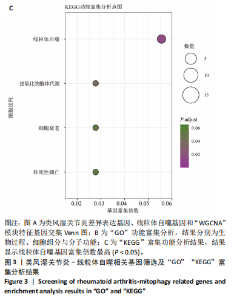
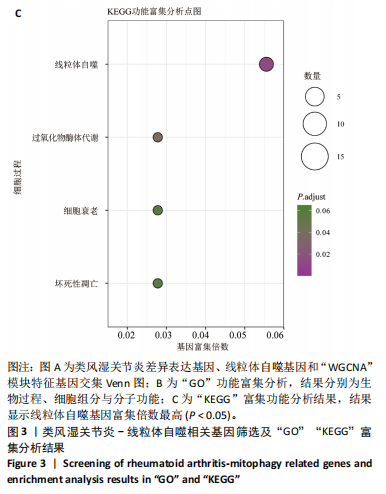
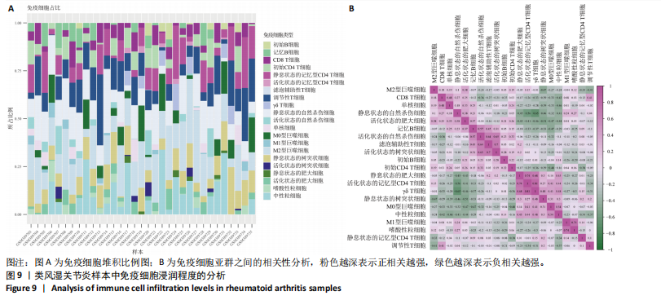
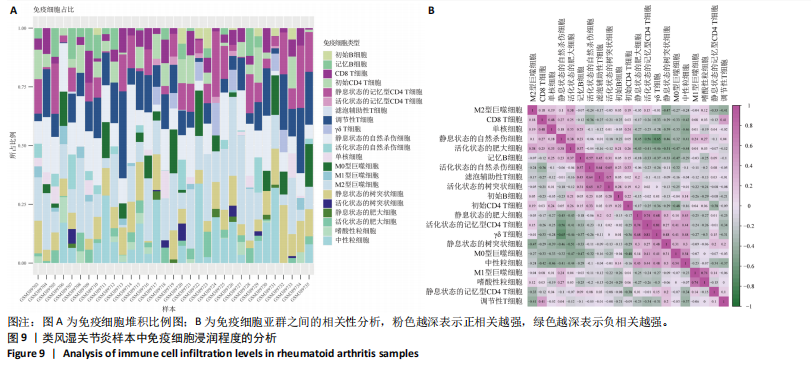
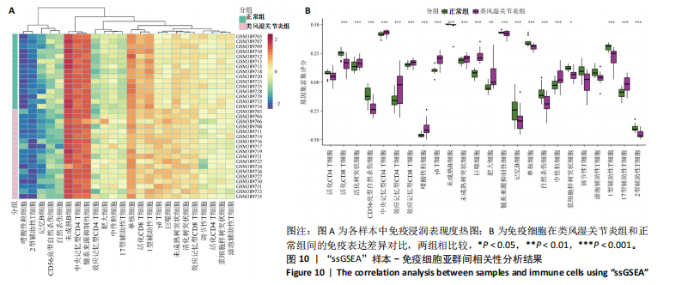
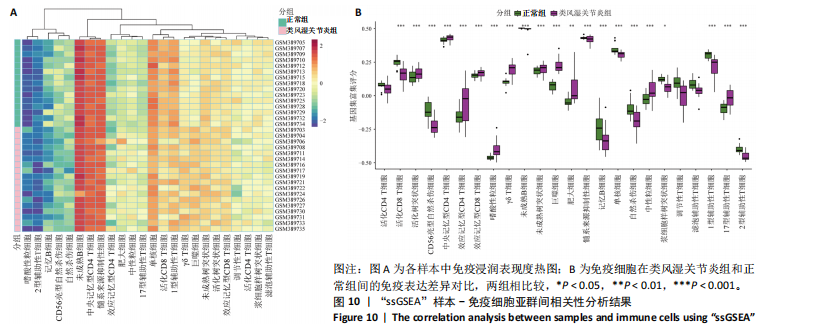
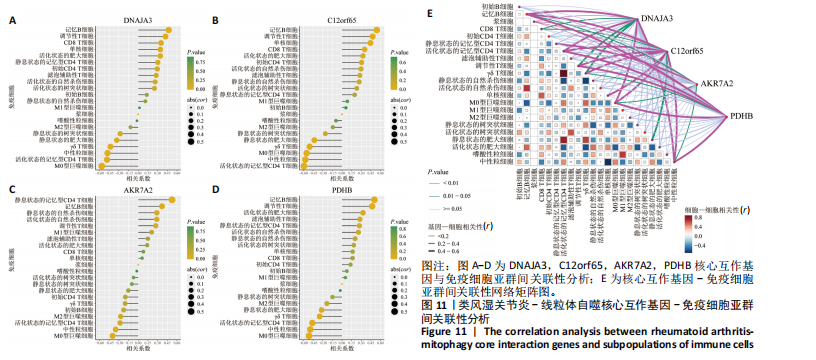
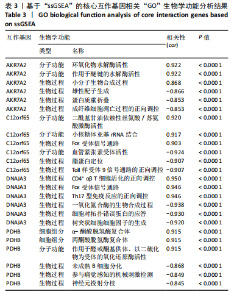
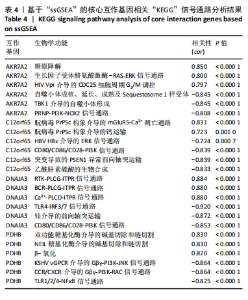
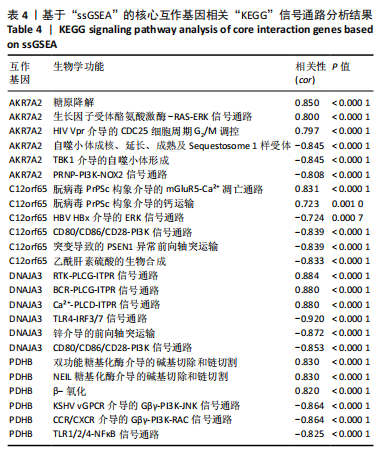
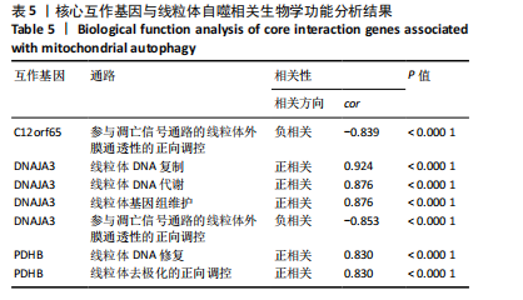
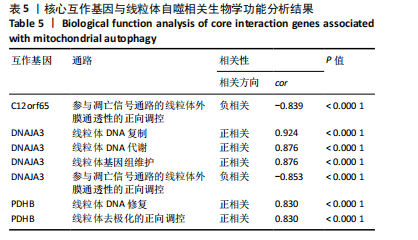
2.9 “CIBERSORT”免疫浸润分析结果 通过“CIBERSORT”算法,利用“MsigDB”数据库的22种免疫细胞“gmt”数据和类风湿关节炎差异基因表达矩阵数据进行免疫浸润分析,进行免疫细胞堆积比例图可视化(图9A)及免疫细胞相关性热图可视化(图9B),可以看出呈高正相关性的免疫细胞对有活化的记忆性CD4 T细胞与γδT细胞、活化的记忆性CD4 T细胞与静息态肥大细胞、M1型巨噬细胞与嗜酸性粒细胞、γδT细胞与静息态肥大细胞。其中,γδT细胞与静息态肥大细胞、活化的记忆性CD4 T细胞、静息态树突细胞、M0型巨噬细胞、中性粒细胞均具有较高正相关性。呈高负相关性的免疫细胞对有γδT细胞与静息态 NK 细胞、活化的记忆性CD4 T细胞与静息态 NK 细胞、活化的肥大细胞与静息态树突细胞。其中静息态 NK细胞与静息态肥大细胞、活化的记忆性CD4 T细胞、γδT细胞、静息态树突细胞、M0型巨噬细胞、中性粒细胞均有高负相关性。结果表明,在类风湿关节炎线粒体自噬过程中,γδT细胞与活化的记忆性CD4 T细胞的免疫状态较为活跃。 2.10 “ssGSEA”单样本-免疫细胞亚群间关联性分析结果 通过R包“GSVA”相关函数初步分析各样本和免疫细胞亚群间的相关性,鉴定不同样本中相关免疫细胞的浸润丰度,继而进行免疫浸润热图可视化(图10A)及表达差异分组箱式图可视化(图10B)。免疫浸润热图显示类风湿关节炎组γδT细胞、巨噬细胞的表达显著高于正常组,正常组调节性T细胞的表达显著高于类风湿关节炎组。表达差异分组箱式图显示类风湿关节炎组与正常组间活化的CD8 T细胞、CD56亮型自然杀伤细胞、嗜酸性粒细胞、γδT细胞、1型辅助性T细胞、2型辅助性T细胞的表达均具有显著性差异(P < 0.001)。 2.11 类风湿关节炎-线粒体自噬核心互作基因-免疫细胞亚群间关联性分析结果 首先,通过R包“ggcorrplot”分析核心互作基因与免疫细胞亚群间的相关性,根据4个核心互作基因的表达矩阵数据与“CIBERSORT”免疫浸润结果数据,利用Shapiro-Wilk检验与封装好的Spearman相关性分析函数进行相关计算,获得基因与免疫细胞亚群间的关联性数据,继而进行核心互作基因-免疫细胞关联性“棒棒糖”图可视化(图11A-D)。图中所示,DNAJA3、C12orf65、PDHB与记忆B细胞均具有强正相关性,DNAJA3、AKR7A2、PDHB与M0型巨噬细胞均呈强负相关性,此外,C12orf65还与活化的记忆性CD4 T细胞呈强负相关性,AKR7A2与静息态记忆性CD4 T细胞呈强正相关性。 其次,通过R包“linkET”进行汇总性可视化,绘制核心互作基因-免疫细胞亚群间关联性网络矩阵图(图11E),多核心互作基因共同作用时,其中以记忆B细胞、活化的记忆性CD4 T细胞、调节性T细胞、M0型巨噬细胞、中性粒细胞的参与最为重要。同时,在类风湿关节炎线粒体自噬过程中,活化的记忆性CD4 T细胞与γδT细胞、静息态肥大细胞之间的正相关表达作用最强,单核细胞和中性粒细胞之间的负相关表达作用最强。 2.12 基于“ssGSEA”的核心互作基因相关生物学功能分析结果 “ssGSEA”算法下共获得821种与类风湿关节炎-线粒体自噬核心互作基因相关的“GO”生物学功能(|cor| > 0.8,P < 0.001),其中生物过程数量为600种,细胞组分数量为72种,分子功能数量为149种,并筛选出与每个核心互作基因正相关性和负相关性最强的前3位生物学功能进行展示(表3)。同时获得48条与类风湿关节炎-线粒体自噬核心互作基因相关的“KEGG”信号通路(|cor| > 0.8,P < 0.001),同样筛选出与每个核心互作基因正相关性和负相关性最强的前3位信号通路进行展示(表4)。最后,逐条筛选上述全部获得的821种“GO”生物学功能和48条“KEGG”信号通路,发现有6种生物学功能与线粒体自噬密切相关(表5)。"
| [1] DI MATTEO A, BATHON JM, EMERY P. Rheumatoid arthritis. Lancet. 2023;402(10416):2019-2033. [2] LEE EE, SHIN A, LEE J, et al. All-cause and cause-specific mortality of patients with rheumatoid arthritis in Korea: A nation-wide population-based study. Joint Bone Spine. 2022;89(1):105269. [3] ALMUTAIRI K, NOSSENT J, PREEN D, et al. The global prevalence of rheumatoid arthritis: a meta-analysis based on a systematic review. Rheumatol Int. 2021;41(5):863-877. [4] SCOTT IC, WHITTLE R, BAILEY J, et al. Rheumatoid arthritis, psoriatic arthritis, and axial spondyloarthritis epidemiology in England from 2004 to 2020: An observational study using primary care electronic health record data. Lancet Reg Health Eur. 2022;23:100519. [5] WOUTERS F, MAURITS MP, VAN BOHEEMEN L, et al. Determining in which pre-arthritis stage HLA-shared epitope alleles and smoking exert their effect on the development of rheumatoid arthritis. Ann Rheum Dis. 2022;81(1):48-55. [6] NEMTSOVA MV, ZALETAEV DV, BURE IV, et al. Epigenetic Changes in the Pathogenesis of Rheumatoid Arthritis. Front Genet. 2019;10:570. [7] GRAVALLESE EM, FIRESTEIN GS. Rheumatoid Arthritis - Common Origins, Divergent Mechanisms. N Engl J Med. 2023;388(6):529-542. [8] ALIVERNINI S, FIRESTEIN GS, MCINNES IB. The pathogenesis of rheumatoid arthritis. Immunity. 2022;55(12):2255-2270. [9] CLAYTON SA, MACDONALD L, KUROWSKA-STOLARSKA M, et al. Mitochondria as Key Players in the Pathogenesis and Treatment of Rheumatoid Arthritis. Front Immunol. 2021;12:673916. [10] KAN S, DUAN M, LIU Y, et al. Role of Mitochondria in Physiology of Chondrocytes and Diseases of Osteoarthritis and Rheumatoid Arthritis. Cartilage. 2021;13(2_suppl):1102S-1121S. [11] ASHRAFI G, SCHWARZ TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31-42. [12] ONISHI M, YAMANO K, SATO M, et al. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. [13] ANSARI MY, KHAN NM, AHMAD I, et al. Parkin clearance of dysfunctional mitochondria regulates ROS levels and increases survival of human chondrocytes. Osteoarthritis Cartilage. 2018;26(8):1087-1097. [14] GOODSWEN SJ, BARRATT JLN, KENNEDY PJ, et al. Machine learning and applications in microbiology. FEMS Microbiol Rev. 2021;45(5):fuab015. [15] 李晨曦,石婕,魏巍,等.颞下颌关节PVNS和RA共病机制及潜在治疗靶点的初步研究[J].生物信息学,2025,23(1):71-80. [16] RATH S, SHARMA R, GUPTA R, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49(D1):D1541-D1547. [17] CASTANZA AS, RECLA JM, EBY D, et al. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat Methods. 2023;20(11):1619-1620. [18] QIAN H, DENG C, CHEN S, et al. Targeting pathogenic fibroblast-like synoviocyte subsets in rheumatoid arthritis. Arthritis Res Ther. 2024; 26(1):103. [19] MANKIA K, D’AGOSTINO MA, ROWBOTHAM E, et al. MRI inflammation of the hand interosseous tendons occurs in anti-CCP-positive at-risk individuals and may precede the development of clinical synovitis. Ann Rheum Dis. 2019;78(6):781-786. [20] ALETAHA D, SMOLEN JS. Diagnosis and Management of Rheumatoid Arthritis: A Review. JAMA. 2018;320(13):1360-1372. [21] SHI Y, ZHOU M, CHANG C, et al. Advancing precision rheumatology: applications of machine learning for rheumatoid arthritis management. Front Immunol. 2024;15:1409555. [22] NAGY G, ROODENRIJS NMT, WELSING PM, et al. EULAR definition of difficult-to-treat rheumatoid arthritis. Ann Rheum Dis. 2021;80(1):31-35. [23] DETER RL, DE DUVE C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33(2):437-449. [24] MIZUSHIMA N, KOMATSU M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728-741. [25] LEMASTERS JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8(1):3-5. [26] WANG Y, LIU N, LU B. Mechanisms and roles of mitophagy in neurodegenerative diseases. CNS Neurosci Ther. 2019;25(7):859-875. [27] WANG S, DENG Z, MA Y, et al. The Role of Autophagy and Mitophagy in Bone Metabolic Disorders. Int J Biol Sci. 2020;16(14):2675-2691. [28] HUANG T, WANG Y, YU Z, et al. Effect of mitophagy in the formation of osteomorphs derived from osteoclasts. iScience. 2023;26(5):106682. [29] ZHAO RZ, JIANG S, ZHANG L, et al. Mitochondrial electron transport chain, ROS generation and uncoupling (Review). Int J Mol Med. 2019; 44(1):3-15. [30] WANG S, LONG H, HOU L, et al. The mitophagy pathway and its implications in human diseases. Signal Transduct Target Ther. 2023;8(1):304. [31] YANG YD, LI ZX, HU XM, et al. Insight into Crosstalk Between Mitophagy and Apoptosis/Necroptosis: Mechanisms and Clinical Applications in Ischemic Stroke. Curr Med Sci. 2022;42(2):237-248. [32] LIU Y, LUO X, CHEN Y, et al. Heterogeneous ferroptosis susceptibility of macrophages caused by focal iron overload exacerbates rheumatoid arthritis. Redox Biol. 2024;69:103008. [33] HAN J, LUO J, WANG C, et al. Roles and mechanisms of copper homeostasis and cuproptosis in osteoarticular diseases. Biomed Pharmacother. 2024;174:116570. [34] SAYSON SL, FAN JN, KU CL, et al. DNAJA3 regulates B cell development and immune function. Biomed J. 2024;47(2):100628. [35] MALETZKO A, KEY J, WITTIG I, et al. Increased presence of nuclear DNAJA3 and upregulation of cytosolic STAT1 and of nucleic acid sensors trigger innate immunity in the ClpP-null mouse. Neurogenetics. 2021;22(4):297-312. [36] SHIN CS, MENG S, GARBIS SD, et al. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat Commun. 2021;12(1):265. [37] WU C, TAN S, LIU L, et al. Transcriptome-wide association study identifies susceptibility genes for rheumatoid arthritis. Arthritis Res Ther. 2021;23(1):38. [38] JIANG M, LIU K, LU S, et al. Verification of cuproptosis-related diagnostic model associated with immune infiltration in rheumatoid arthritis. Front Endocrinol (Lausanne). 2023;14:1204926. [39] DESAI N, YANG H, CHANDRASEKARAN V, et al Elongational stalling activates mitoribosome-associated quality control. Science. 2020; 370(6520):1105-1110. [40] BREDA CNS, DAVANZO GG, BASSO PJ, et al. Mitochondria as central hub of the immune system. Redox Biol. 2019;26:101255. [41] MA C, WANG J, HONG F, et al. Mitochondrial Dysfunction in Rheumatoid Arthritis. Biomolecules. 2022;12(9):1216. |
| [1] | Zhang Yibo, Lu Jianqi, Mao Meiling, Pang Yan, Dong Li, Yang Shangbing, Xiao Xiang. Exploring the causal relationship between rheumatoid arthritis and coronary atherosclerosis: a Mendel randomized study involving serum metabolites and inflammatory factors [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-9. |
| [2] | Li Jiagen, Chen Yueping, Huang Keqi, Chen Shangtong, Huang Chuanhong. The construction and validation of a prediction model based on multiple machine learning algorithms and the immunomodulatory analysis of rheumatoid arthritis from the perspective of mitophagy [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(在线): 1-15. |
| [3] | Han Haihui, Ran Lei, Meng Xiaohui, Xin Pengfei, Xiang Zheng, Bian Yanqin, Shi Qi, Xiao Lianbo. Targeting fibroblast growth factor receptor 1 signaling to improve bone destruction in rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(9): 1905-1912. |
| [4] | Wang Yuru, Li Siyuan, Xu Ye, Zhang Yumeng, Liu Yang, Hao Huiqin. Effects of wogonin on joint inflammation in collagen-induced arthritis rats via the endoplasmic reticulum stress pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1026-1035. |
| [5] | Liu Yani, Yang Jinghuan, Lu Huihui, Yi Yufang, Li Zhixiang, Ou Yangfu, Wu Jingli, Wei Bing . Screening of biomarkers for fibromyalgia syndrome and analysis of immune infiltration [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 1091-1100. |
| [6] | Han Haihui, Meng Xiaohu, Xu Bo, Ran Le, Shi Qi, Xiao Lianbo. Effect of fibroblast growth factor receptor 1 inhibitor on bone destruction in rats with collagen-induced arthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(5): 968-977. |
| [7] | Yang Dingyan, Yu Zhenqiu, Yang Zhongyu. Machine learning-based analysis of neutrophil-associated potential biomarkers for acute myocardial infarction [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(36): 7909-7920. |
| [8] | Chen Tianxin, , Zhang Zhilong, Zhang Shuai, Gao Yun, Zhu Yuqi, Yang Shengping. Causal relationship between immune cells and bone metabolic diseases: a Mendelian randomization analysis of European populations in international databases [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(29): 6326-6332. |
| [9] | Liu Yuan, Qu Yuan, Wan Yakun, Guo Jingyu, Jiang Ping. Transcriptomic analysis and drug prediction of basement membrane-related genes in different traditional Chinese medicine patterns of rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(25): 5486-5500. |
| [10] | Lin Yixin, Wang Wenyi, Lei Xiaoqing, Ma Dezun, Huang Yanfeng, Fu Changlong, Ye Jinxia. Tougu Xiaotong Capsules for treating arthritis according to the principle of “Same Treatment for Different diseases”: analysis based on integrated pharmacology, molecular docking techniques and molecular dynamics simulation [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(24): 5093-5101. |
| [11] | Zou Shunyi, Chai yuan, Li Kunjian. Involvement of macrophage polarization in osteoarticular diseases: a visual analysis based on SCI-Expanded information [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(24): 5245-5253. |
| [12] | Zhang Yibo, Lu Jianqi, Mao Meiling, Pang Yan, Dong Li, Yang Shangbing, Xiao Xiang. Rheumatoid arthritis and coronary atherosclerosis: data analysis of serum metabolite and inflammatory factor in the European population [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(24): 5263-5271. |
| [13] | Yue Jinru, Zhang Yumin, Liu Jingshu, Bu Yanan, Wu Jingruo, Chen Jia, Wang Jianru. Effects of extracellular vesicles treated with Duhuo Jisheng Decoction on rheumatoid arthritis fibroblast-like synovial cells [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(23): 4915-4926. |
| [14] | Xu Liang, Gulimila·Muhetaer, Ju Bowei, Li Ruoning. Molecular mechanism of allicin-targeted regulation of epidermal growth factor receptor and kynureninase in the treatment of rheumatoid arthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(20): 4205-4214. |
| [15] | Wu Yingkai, Shi Gaolong, Xie Zonggang . Weighted gene co-expression network analysis and machine learning identification of key genes in rheumatoid arthritis synovium [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(2): 294-301. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||