Chinese Journal of Tissue Engineering Research ›› 2025, Vol. 29 ›› Issue (2): 312-321.doi: 10.12307/2025.239
Previous Articles Next Articles
Mechanism by which KRT18 interacts with mRNA and long non-coding RNA to regulate intervertebral disc nucleus pulposus cell injury
Liu Zhongyuan1, Li Yang2, Zhang Zhiwen2
- 1College of Acupuncture and Orthopedics, Hubei University of Chinese Medicine, Wuhan 430061, Hubei Province, China; 2Hospital affiliated to Hubei University of Traditional Chinese Medicine, Hubei Hospital of Traditional Chinese Medicine, Hubei Institute of Traditional Chinese Medicine, Wuhan 430061, Hubei Province, China
-
Received:2023-12-07Accepted:2024-02-19Online:2025-01-18Published:2024-05-24 -
Contact:Zhang Zhiwen, MD, Associate chief physician, Hospital affiliated to Hubei University of Traditional Chinese Medicine, Hubei Hospital of Traditional Chinese Medicine, Hubei Institute of Traditional Chinese Medicine, Wuhan 430061, Hubei Province, China -
About author:Liu Zhongyuan, Master candidate, Physician, College of Acupuncture and Orthopedics, Hubei University of Chinese Medicine, Wuhan 430061, Hubei Province, China -
Supported by:the Natural Science Foundation of Hubei Province, No. 2022CFB406 (to ZZW)
CLC Number:
Cite this article
Liu Zhongyuan, Li Yang, Zhang Zhiwen . Mechanism by which KRT18 interacts with mRNA and long non-coding RNA to regulate intervertebral disc nucleus pulposus cell injury[J]. Chinese Journal of Tissue Engineering Research, 2025, 29(2): 312-321.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
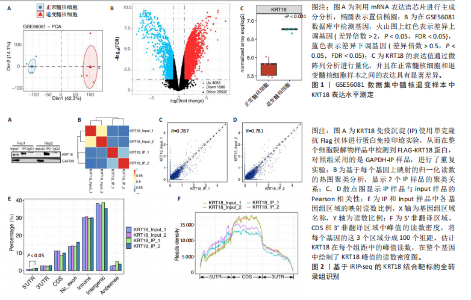
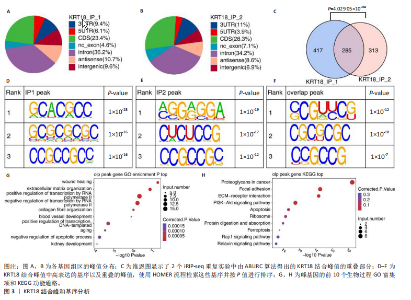
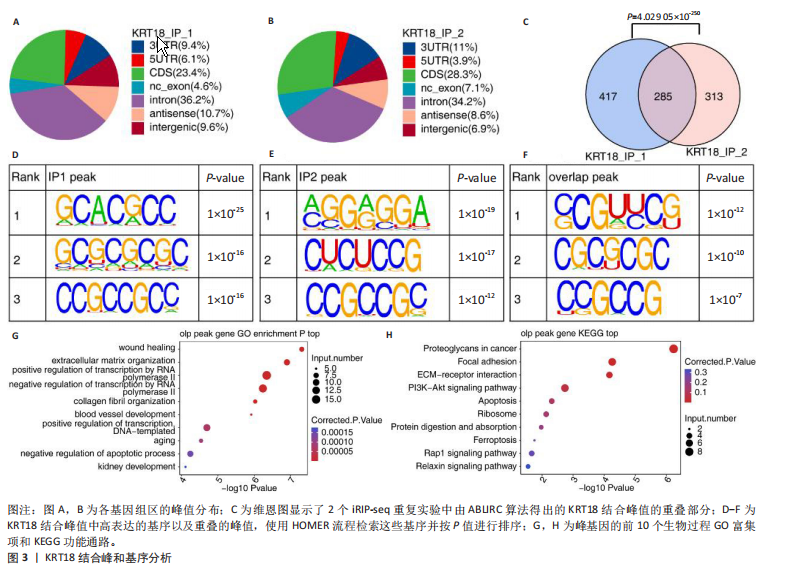
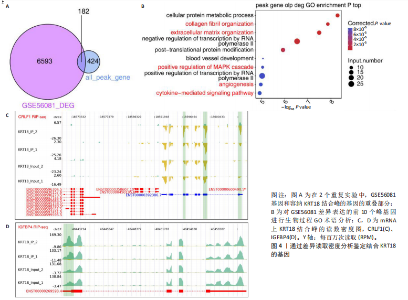
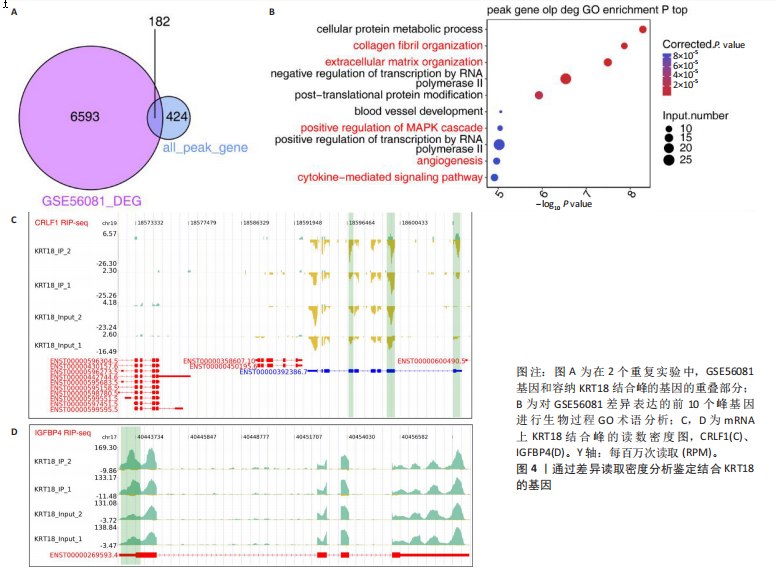
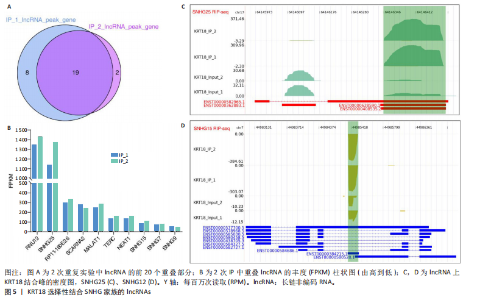
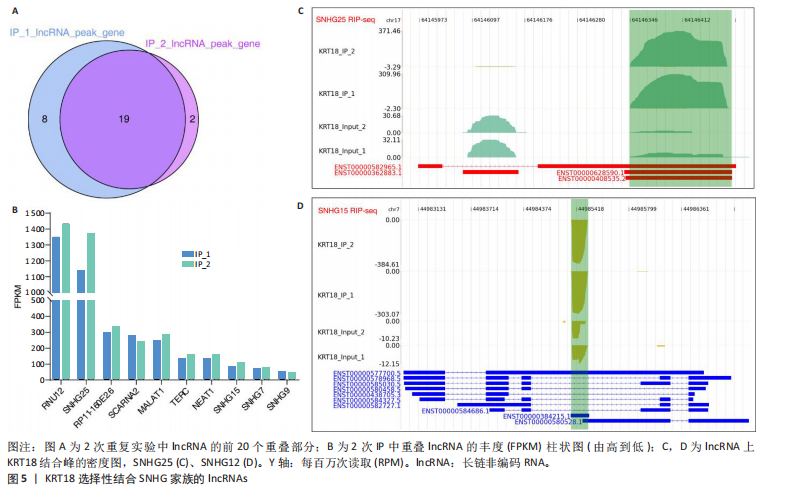
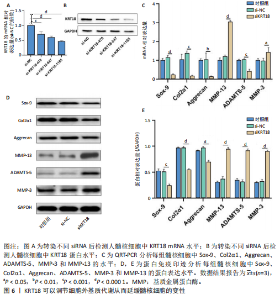
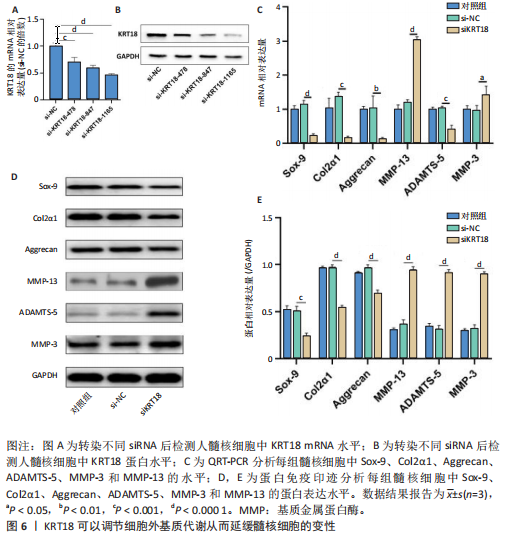
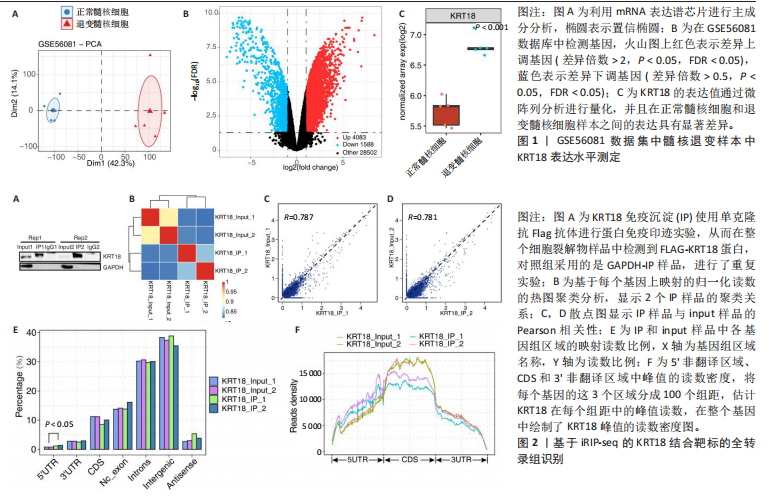
2.1 KRT18在退变髓核组织中表达并广泛影响转录组图谱 现有研究表明,椎间盘退变的发病机制包括细胞外基质相关蛋白、炎症因子和细胞因子的失调等[22-24]。角蛋白家族尤其是KRT18被认为是单细胞水平上椎间盘的潜在生物标志物,但其功能尚不清楚[25]。为了进一步研究KRT18对椎间盘退行性病变的潜在影响,分析了GSE56081转录组数据集,其中包含5个退变髓核样本和5个健康髓核样本[26]。在得到所有检测到的基因水平变化后,通过对退变髓核和健康髓核样本中的所有基因进行主成分分析,可以清晰地分离出这两组(图1A),表明了髓核转录组图谱的整体变化。MA图展示了与椎间盘退行性病变相关的差异表达基因,表明上调的差异表达基因明显多于下调的(图1B)。在4 083个上调的差异表达基因中,通过微阵列分析检测到在退变髓核组织中KRT18存在明显较高的表达(图1C),暗示KRT18表达水平的变化能够密切调控影响椎间盘退行性病变的发生。 2.2 KRT18-RNA 相互作用图揭示了 KRT18 与成熟 mRNA 的结合能力 KRT18在先前的一项研究中被证实是一种新的RNA结合蛋白,暗示了其在椎间盘退变中的潜在功能[27]。为了深入研究KRT18在椎间盘退行性病变中可能发挥的功能,采用了一种改进的RNA免疫沉淀和测序方法(iRip-seq)来获得髓核细胞内与KRT18结合的RNA的全面基因组图谱。为此在iRIP-seq 过程中使用了Flag标记的 KRT18并进行2次独立的重复试验。蛋白免疫印迹实验验证了Flag-KRT18蛋白在总细胞裂解物和 Flag免疫沉淀 (IP) 样品中存在,而在IgG对照的IP部分中则不存在(图2A),从而验证了iRIP过程的成功执行。将低质量的测序读数筛除后与人类基因组进行比对,随后根据对所有4个样本(2个IP样本和2个input样本)在整个转录组中映射读数分析得到,2个KRT18_IP样本展现出与input样本明显不同的聚类模式(图2B)。此外,IP和input样本对中每个检测到基因表达水平的相关图(以FPKM为单位)显示了KRT18_IP样本中特定基因的某些mRNA的偏倚(图2C,D),这些结果证明 iRIP-seq 实验已成功进行。KRT18_IP和input样本中映射读数的基因组位置显示在5’非翻译区域KRT18相关读数存在富集(图2E)。对成熟mRNA的3个区域进行标准化读取密度分析进一步确认了KRT18在5’非翻译区域的富集(图2F)。5’非翻译是mRNA的起始区域,通常包含有调控转录和翻译的序列元件。基因在这个区域富集可能意味着它在调控转录和翻译过程中起着重要的作用,可能与mRNA的稳定性、翻译效率或其他生物学功能相关联。因此,这种富集可能表明KRT18与mRNA之间存在着良好的结合关系。 2.3 KRT18 结合峰富含CCG和CGC的基序 从iRip-seq读数中提取KRT18结合峰,通过ABLIRC流程进行分析[18],以研究髓核细胞内KRT18对与其结合的mRNA的影响。这些峰值的基因组位置主要存在于3’非翻译区域、内含子和编码序列(图 3A,B)。进一步分析重叠峰发现在2个重复样本之间共有285个峰共享基因组位置(图3C,P=4.029 05×10-250,超几何检验)。为了找出KRT18 峰中富集的潜在序列基序,采用了 HOMER 算法,显示共有序列CCG和CGC在两次IP实验识别的峰中都明显更多(图 3D,E),这两个基序在共享峰中排名前三,P值具有差异显著性(图3F)。说明与KRT18相互作用的mRNA在转录和翻译调控中具有重要的功能。随后对与KRT18相互作用的mRNA进行了富集功能分析,GO分析得到一些与椎间盘退行性病变强相关的生物过程通路(图3G),包括细胞外基质组织[28-29]、胶原纤维组织[30]、DNA转录调控、衰老以及凋亡的负调节;此外,KEGG分析显示其在黏附斑和细胞外基质-受体相互作用途径中存在显著富集(图3H);同时这些基因在凋亡和松弛素信号通路中也被发现富集[31]。这些发现表明KRT18在相关mRNA的转录和选择性剪接过程中具有重要调控作用,密切影响椎间盘退行性病变的发病机制。 2.4 KRT18结合mRNA的基因分析 为了进一步阐明与KRT18结合的mRNA在椎间盘退行性病变的潜在活性功能,将GSE56081数据集的差异表达基因与2次iRIP-seq中检测到的KRT18结合的基因峰群进行交叉,得到了182个重叠基因(图4A),对这182个基因进行GO功能分析,分析结果显示高度富集的通路有细胞外基质组织、胶原纤维组织、MAPK级联的正向调节、细胞因子介导的信号传导途径等(图4B)。此外结果发现KRT18能够良好的结合到CRLF1和IGFBP4的5’非翻译区域(图4C,D),其中CRLF1表达升高是黄韧带肥厚的主要反应[32];而IGFBP4在调节细胞生长、增殖和凋亡中发挥重要作用[33]。因此推测,与KRT18结合的mRNA能够影响调控多种生物功能,并调节细胞外基质代谢,在椎间盘退行性病变的机制中发挥重要作用。 2.5 KRT18结合lncRNA的基因分析 LncRNA作为调节分子在许多生物过程和疾病包括椎间盘退行性病变中发挥着至关重要的作用[34-35]。对KRT18 iRIP-seq 数据的分析显示,两次实验中4.6%和7.1% 的峰位于非编码RNA的外显子区域区域内,该区域包含lncRNA。因此,根据所识别的基因峰群搜索与KRT18相互作用的lncRNA,将2次重复实验中筛选得到的lncRNA(包括lincRNA和反义RNA)交叉获得19个重叠的lncRNA(图5A)。在这些重叠的lncRNA中,RNU12和SNHG25始终表现出高丰度(图5B),表明KRT18能够优先与它们相互作用。将input样本中高表达的lncRNA筛除后发现KRT18与SNHG25和SNHG12能够特异性结合,进一步可视化了这2个 lncRNA 的读数分布和结合峰(图 5C,D),发现KRT18与其能够良好的结合。先前的研究表明,SNHG25能够调控细胞增殖及细胞凋亡[36],同时SNHG12对炎症因子具有良好的调控作用[37]。这些发现表明KRT18与功能性 lncRNA存在较密切的相互作用,能够通过调节髓核细胞环境及炎症反应影响椎间盘退行性病变的发展。 2.6 KRT18调节细胞外基质代谢,延缓髓核细胞变性 为了进一步阐明KRT18对人髓核细胞外基质的调节影响,在人髓核细胞中使用siKRT18敲低KRT18的表达。KRT18敲低的效果通过qRT-PCR和蛋白免疫印迹进行验证,其中si-KRT18-1165表现出最明显的敲低效果(图6A,B)。在细胞外基质方面,Sox-9、Col2α1、Aggrecan、MMP-13、ADAMTS-5和MMP-3等标志物提示椎间盘退变。qRT-PCR和蛋白免疫印迹结果显示KRT18敲低后MMP-3、MMP-13和ADAMTS-5水平显著升高,同时Sox-9、Col2α1和Aggrecan显著降低(图6C-E)。这些发现表明 KRT18 可能通过调节细胞外基质代谢在改善椎间盘退变中发挥关键作用。"
| [1] CIEZA A, CAUSEY K, KAMENOV K, et al.Global estimates of the need for rehabilitation based on the Global Burden of Disease study 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2021;396(10267):2006-2017. [2] MAHER C, UNDERWOOD M, BUCHBINDER R.Non-specific low back pain. Lancet. 2017; 389(10070):736-747. [3] FRANCISCO V, PINO J, GONZÁLEZ-GAY MÁ, et al. A new immunometabolic perspective of intervertebral disc degeneration. Nat Rev Rheumatol. 2022;18(1):47-60. [4] 王乙.TGF-β通路在椎间盘退变分子生物学进程中作用机制的研究进展[J].吉林大学学报(医学版),2020,46(5):1105-1110. [5] XU C, LUO S, WEI L, et al. Integrated transcriptome and proteome analyses identify novel regulatory network of nucleus pulposus cells in intervertebral disc degeneration. BMC Med Genomics. 2021;14(1):40. [6] GEBAUER F, SCHWARZL T, VALCÁRCEL J, et al.RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185-198. [7] 曾如馨,陈鹏.RNA结合蛋白的组学解析与功能探索[J].化学学报,2024,82(1):53-61. [8] KELAINI S, CHAN C, CORNELIUS VA, et al. RNA-Binding Proteins Hold Key Roles in Function, Dysfunction, and Disease.Biology (Basel). 2021;10(5):366. [9] SHAO Z, NI L, HU S,et al. RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration. Cell Prolif. 2021;54(2):e12975. [10] SHAO Z, TU Z, SHI Y, et al. RNA-Binding Protein HuR Suppresses Inflammation and Promotes Extracellular Matrix Homeostasis via NKRF in Intervertebral Disc Degeneration. Front Cell Dev Biol. 2020;8: 611234. [11] PAN H, STRICKLAND A, MADHU V, et al. RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1α signaling in nucleus pulposus cells. Matrix Biol. 2019; 77:23-40. [12] WANG D, SHANG Q, MAO J, et al. Phosphorylation of KRT8 (keratin 8) by excessive mechanical load-activated PKN (protein kinase N) impairs autophagosome initiation and contributes to disc degeneration. Autophagy. 2023;19(9): 2485-2503. [13] MURIEL JM, O’NEILL A, KERR JP, et al. Keratin 18 is an integral part of the intermediate filament network in murine skeletal muscle. Am J Physiol Cell Physiol. 2020;318(1):C215-C224. [14] BAEK A, SON S, BAEK YM, et al. KRT8 (keratin 8) attenuates necrotic cell death by facilitating mitochondrial fission-mediated mitophagy through interaction with PLEC (plectin). Autophagy. 2021;17(12):3939-3956. [15] WANG YXJ. Several concerns on grading lumbar disc degeneration on MR image with Pfirrmann criteria. J Orthop Translat. 2022:32:101-102. [16] 赵延辉,陈少康,翟丽维,等.RNA-seq数据差异表达分析流程比较[J].中国农业大学学报,2023,28(6):153-159. [17] UREN PJ, BAHRAMI-SAMANI E, BURNS SC, et al. Site identification in high-throughput RNA–protein interaction data. Bioinformatics. 2012;28(23):3013-3020. [18] XIA H, CHEN D, WU Q, et al. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta Gene Regul Mech. 2017;1860(9):911-921. [19] ANDREACE F, LECHAT P, DUFRESNE Y, et al. Comparing methods for constructing and representing human pangenome graphs. Genome Biol. 2023;24(1):274. [20] XIE C, MAO X, HUANG J, et al.KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue): W316-322. [21] GOU J.On dependence assumption in p-value based multiple test procedures. J Biopharm Stat. 2023;33(5):596-610. [22] 彭兵,杜立龙,张同星,等.椎间盘源性腰痛的发病机制与治疗进展[J].中国矫形外科杂志,2023,31(16):1488-1492. [23] JHA R, BERNSTOCK JD, CHALIF JI, et al.Updates on Pathophysiology of Discogenic Back Pain. J Clin Med. 2023;12(21):6907. [24] KANG L, ZHANG H, JIA C, et al.Epigenetic modifications of inflammation in intervertebral disc degeneration. Ageing Res Rev. 2023;87:101902. [25] CALISKAN A, CROUCH SAW, GIDDINS S, et al. Progeria and Aging-Omics Based Comparative Analysis. Biomedicines. 2022; 10(10):2440. [26] LIU XW, XU HW, YI YY, et al. Role of ferroptosis and immune infiltration in intervertebral disc degeneration: novel insights from bioinformatics analyses. Front Cell Dev Biol. 2023;11:1170758. [27] 张立存,赵继荣,徐兵,等.中医药干预治疗椎间盘退行性变基因学研究进展[J].中国老年学杂志,2022,42(9):2288-2292. [28] 赵继荣,杨正汉,马俊飞,等.中医药干预基质金属蛋白酶表达治疗椎间盘退变研究进展[J].中国实验方剂学杂志,2023, 29(5):272-282. [29] KAKUTANI K, YURUBE T, AN HS, et al. Cytokine Inhibitors Upregulate Extracellular Matrix Anabolism of Human Intervertebral Discs under Alginate Beads and Alginate-Embedded Explant Cultures. Int J Mol Sci. 2023;24(15):12336. [30] 王志剑.腰椎间盘胶原酶溶解术临床应用中国专家共识[J].中国疼痛医学杂志, 2022,28(2):81-85. [31] XU J, WAN S, CHEN W, et al. Relaxin inhibits 177Lu-EDTMP associated cell death in osteosarcoma cells through notch-1 pathway. Acta Pharm. 2022;72(4):575-585. [32] ZHENG Z, AO X, LI P, et al. CRLF1 is a key regulator in the ligamentum flavum hypertrophy. Front Cell Dev Biol. 2020: 8:858. [33] VITALI E, GRASSO A, SCHIAVONE ML, et al. The direct impact of pegvisomant on osteoblast functions and bone development. J Endocrinol Invest. 2023 Dec 30. doi: 10.1007/s40618-023-02281-3. [34] 王亮,张虎林,汪小敏,等.LncRNA在椎间盘退变中的作用机制及临床应用前景[J].中国临床解剖学杂志,2023,41(2): 245-247. [35] WU Y, LI S, SHEN J, et al. Nucleus pulposus related lncRNA and mRNA expression profiles in intervertebral disc degeneration. Genomics. 2023;115(2):110570. [36] WAN N, LIU Q, SHI J, et al. LncRNA SNHG25 predicts poor prognosis and promotes progression in osteosarcoma via the miR-497-5p/SOX4 axis. Comb Chem High Throughput Screen. 2023 Jun 2. doi: 10.2174/1386207326666230602122618. [37] BUZZATTO-LEITE I, AFONSO J, SILVA-VIGNATO B, et al. Differential gene co-expression network analyses reveal novel molecules associated with transcriptional dysregulation of key biological processes in osteoarthritis knee cartilage. Osteoarthr Cartil Open. 2022;4(4):100316. [38] 梁松林,李志超,高尚,等.中药单体促髓核细胞自噬缓解椎间盘退变的研究进展[J/OL].中华中医药学刊:1-12[2024-01-21]. https://link.cnki.net/urlid/21.1546.r.20230824.1334.018 [39] 姚智,魏梦诚,刘伟军,等.腺苷受体2A调控神经肽Y抑制髓核细胞凋亡及基质降解的机制研究[J].中华实验外科杂志, 2023,40(12):2525-2528. [40] MURAKAMI S, JAFFREY SR. Hidden codes in mRNA: Control of gene expression by m6A. Mol Cell. 2022;82(12):2236-2251. [41] CHEN Y, HUA Q, WAN H, et al. Long Noncoding RNA SLC20A1-1 Induces Nucleus Pulposus Apoptosis by Sponging miR-146a-5p. Genet Test Mol Biomarkers. 2022;26(3):127-132. [42] LAI H, LI Y, ZHANG H, et al. exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022;50(D1): D118-D128. [43] WU T, CHENG AY, ZHANG Y, et al. KARR-seq reveals cellular higher-order RNA structures and RNA-RNA interactions. Nat Biotechnol. 2024. doi: 10.1038/s41587-023-02109-8. [44] CUI J, MA Q, ZHANG C, et al. Keratin 18 Depletion as a Possible Mechanism for the Induction of Apoptosis and Ferroptosis in the Rat Hippocampus After Hypobaric Hypoxia. Neuroscience. 2023;513:64-75. [45] BLANC V, MOLITOR EA, DAVIDSON NO. Protocol to isolate RBP-mRNA complexes using RNA-CLIP and examine target mRNAs. STAR Protoc. 2023;4(2):102313. [46] CHEN B, XU X, LIN DD, et al. KRT18 Modulates Alternative Splicing of Genes Involved in Proliferation and Apoptosis Processes in Both Gastric Cancer Cells and Clinical Samples. Front Genet. 2021; 12:635429. [47] WANG JX, ZHAO X, XU SQ.Screening Key lncRNAs of Ankylosing Spondylitis Using Bioinformatics Analysis. J Inflamm Res. 2022;15:6087-6096. [48] NICHOLSON CO, FRIEDERSDORF M, KEENE JD. Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. Rna. 2017;23(1):32-46. [49] DIEZ-HERMANO S, MEJIAS A, SANCHEZ D, et al. Control of the neuroprotective Lipocalin Apolipoprotein D expression by alternative promoter regions and differentially expressed mRNA 5’ UTR variants. PLoS One. 2020;15(6):e0234857. [50] LI C, SUN C, MAHAPATRA KD, et al.Long noncoding RNA plasmacytoma variant translocation 1 is overexpressed in cutaneous squamous cell carcinoma and exon 2 is critical for its oncogenicity. Br J Dermatol. 2024;190(3):415-426. [51] CHENG Y, QIN K, HUANG N, et al.Cytokeratin 18 regulates the transcription and alternative splicing of apoptotic‑related genes and pathways in HeLa cells. Oncol Rep. 2019;42(1):301-312. [52] PAN L, E T, XU C, et al. The apoptotic effects of soybean agglutinin were induced through three different signal pathways by down-regulating cytoskeleton proteins in IPEC-J2 cells. Sci Rep. 2023;13(1):5753. [53] LIU Y, XU B, LIU M, et al.Long non-coding RNA SNHG25 promotes epithelial ovarian cancer progression by up-regulating COMP. J Cancer. 2021;12(6):1660-1668. [54] GHAFOURI-FARD S, SHOOREI H, HUSSEN BM, et al. LncRNA SNHG12: A budding star in human diseases. Pathol Res Pract. 2023;251:154897. |
| [1] | Wu Wangxiang, Ran Dongcheng, Xu Jiamu, Xu Jiafu, Chen Jingjing, Wang Chunqing. Long noncoding RNAs related to osteoporosis: current research status and developmental trends [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(29): 6360-6368. |
| [2] | Wang Runzheng, Fu Su, Dong Chao, Li Dongzhe, Wang Yongkui. Relationship between bone mineral density and lumbar disc degeneration in middle-aged and elderly men and postmenopausal women [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(24): 5079-5085. |
| [3] | Zhang Yanfeng, Zhang Huimin, He Xiang, Zheng Yuping. Effects of long non-coding RNA nuclear enriched abundant transcript 1 on the proliferation, apoptosis and migration of keloid fibroblasts [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(2): 347-354. |
| [4] | Zheng Shanbin, Xia Tianwei, Sun Jiahao, Chen Zhiyuan, Cao Xun, Zhang Chao, Shen Jirong . Relationship between long non-coding RNA and osteoarthritis [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(11): 2357-2367. |
| [5] | Qin Hao, Kang Teng, Liu Gang. Long non-coding RNA directly or indirectly affects osteoporosis through p38MAPK signaling pathway [J]. Chinese Journal of Tissue Engineering Research, 2025, 29(1): 175-184. |
| [6] | Lin Zhanying, Lin Ziyun, Huang Liuyan, Zhang Wenxi, Zuo Changqing. Overexpression of long non-coding RNA Gm16104 affects osteogenic differentiation of C3H10T1/2 mesenchymal stem cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(31): 4964-4969. |
| [7] | Hu Lingli, Li Na, Li Jingyang, Zhang Eryun, Chen Yu, Gu Ying. Role of non-coding RNA and exosomes in pathogenesis of gestational diabetes mellitus and their early diagnostic value [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(31): 5070-5077. |
| [8] | Liu Ying, Liu Yalei, Liu Yu. Screening and analysis of differentially expressed long non-coding RNAs in adriamycin-induced myocardial injury antagonized with daisy leaf gentianone [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(26): 4121-4128. |
| [9] | Wan Jun, Bai Yanjie, Wang Yan, Chen Shuying, Chen Limin, Xiao Yuqian, Sun Kexin. Mechanism of action and related signaling pathways of long non-coding RNAs in neuroimmuno-inflammatory response after ischemic stroke [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(20): 3265-3271. |
| [10] | Tong Tong, Liu Chunyan, Liu Bing, Zhao Fei. Long non-coding RNA and periodontitis [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(14): 2267-2273. |
| [11] | Fan Zhihong, Zhang Xian, Li Chao. Wnt signaling pathway in intervertebral disc degeneration [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(12): 1950-1955. |
| [12] | Wang Xianfeng, Wang Kun, Sun Han, Sun Xiaoliang, Yan Litao. Mechanism underlying exosomal lncRNA H19 derived from umbilical cord mesenchymal stem cells promotes cartilage injury repair [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 20-25. |
| [13] | Zhou Minghua, Hu Xiaoyu. LncRNA SNHG4 regulates miR-152-3p during osteoblastic differentiation of periodontal ligament stem cells [J]. Chinese Journal of Tissue Engineering Research, 2024, 28(1): 38-43. |
| [14] | Guo Shuhui, Yang Ye, Jiang Yangyang, Xu Jianwen. Screening and validation of neurogenic bladder miRNA-mRNA regulatory network [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(在线): 1-8. |
| [15] | Hu Xinming, Qiao Yanhua, Wang Xiaofan, Li Linyu, Zhao Bing. Mechanism of long non-coding RNA plasmacytoma variant translocation 1 involved in pelvic organ prolapse [J]. Chinese Journal of Tissue Engineering Research, 2023, 27(5): 669-675. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||